HCA Data Portal Platform Updates
June 16, 2025
New projects (5):
- Mapping human haematopoietic stem cells from haemogenic endothelium to birth.
- Molecular phenotyping reveals the identity of Barrett’s esophagus and its malignant transition
- Single-Cell Characterization of Malignant Phenotypes and Developmental Trajectories of Adrenal Neuroblastoma
- Single-cell roadmap of human gonadal development
- Transcriptomic and spatial dissection of human ex vivo right atrial tissue reveals proinflammatory microvascular changes in ischemic heart disease
Projects with updated metadata (1):
June 2, 2025
New projects (2):
- An atlas of transcribed enhancers across helper T cell diversity for decoding human diseases
- Single-Cell Transcriptomic Comparison of Human Fetal Retina, hPSC-Derived Retinal Organoids, and Long-Term Retinal Cultures
Projects with updated files (5):
- Capturing human trophoblast development with naive pluripotent stem cells in vitro
- Integrated scRNA-Seq Identifies Human Postnatal Thymus Seeding Progenitors and Regulatory Dynamics of Differentiating Immature Thymocytes
- Microglia Require CD4 T Cells to Complete the Fetal-to-Adult Transition.
- Single-cell RNA-sequencing reveals the landscape of cervical cancer heterogeneity
- Tabula Muris: Transcriptomic characterization of 20 organs and tissues from Mus musculus at single cell resolution
May 16, 2025
New projects (5):
- An integrated single-cell reference atlas of the human endometrium
- Cellular heterogeneity and dynamics of the human uterus in healthy premenopausal women
- Single-cell RNA-sequencing of skin, fresh blood, and cultured peripheral blood mononuclear cells from a patient with drug-induced hypersensitivity syndrome and healthy volunteers
- Single-cell atlas of the human neonatal small intestine affected by necrotizing enterocolitis
- Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex
Projects with updated files (7):
- AIDA
- Molecular transitions in early progenitors during human cord blood hematopoiesis
- Single cell RNAseq analysis of the developmental trajectory of iPSC-derived tenocytes
- Single-cell RNA Sequencing of human microglia from post mortem Alzheimers Disease CNS tissue.
- Single-cell survey of human lymphatics unveils marked endothelial cell heterogeneity and mechanisms of homing for neutrophils
- The Human and Mouse Enteric Nervous System at Single-Cell Resolution.
- Transcriptome Landscape of Human Folliculogenesis Reveals Oocytes and Granulosa Cells Interactions
April 16, 2025
New projects (5):
- A single-cell atlas of human and mouse white adipose tissue
- Single-cell landscape of functionally cured chronic hepatitis B patients reveals activation of innate and altered CD4-CTL-driven adaptive immunity
- Single-nucleus RNA-seq in the post-mortem brain in major depressive disorder.
- Spatial transcriptomics of healthy and fibrotic human liver at single-cell resolution
- Type I interferon autoantibodies are associated with systemic immune alterations in patients with COVID-19
Febraury 24, 2025
New projects (8):
- A human omentum-specific mesothelial-like stromal population inhibits adipogenesis through IGFBP2 secretion
- A single‐cell RNA expression atlas of normal, preneoplastic and tumorigenic states in the human breast
- An integrated single cell and spatial transcriptomic map of human white adipose tissue
- Early-life inflammation primes a T helper 2 cell-fibroblast niche in skin
- Preneoplastic stromal cells promote BRCA1-mediated breast tumorigenesis
- Single cell full-length transcriptome of human subcutaneous adipose tissue reveals unique and heterogeneous cell populations
- Spatial mapping reveals human adipocyte subpopulations with distinct sensitivities to insulin
- The Dynamic Transcriptional Cell Atlas of Testis Development During Human Puberty
Updated Projects (2):
- A single-cell transcriptome atlas of human early embryogenesis
- Assessing Cellular and Transcriptional Diversity of Ileal Mucosa Among Treatment-Naïve and Treated Crohn’s Disease
January 15, 2025
New Raw Data and Metadata (6)
- Concerted changes in the pediatric single-cell intestinal ecosystem before and after anti-TNF blockade
- Human subcutaneous and visceral adipocyte atlases uncover classical and non-classical adipocytes and depot-specific patterns
- Investigating immune and non-immune cell interactions in head and neck tumors by single-cell RNA sequencing
- Mapping hormone-regulated cell-cell interaction networks in the human breast at single-cell resolution
- Multimodal single-cell profiling reveals neuronal vulnerability and pathological cell states in focal cortical dysplasia
- Mural cell composition and functional analysis in the healing process of human gingiva from periodontal intrabony defects
Updated Raw Data and Metadata (5)
- A single-cell transcriptome atlas of human early embryogenesis
- A temporal cortex cell atlas highlights gene expression dynamics during human brain maturation
- Organoid single-cell genomic atlas uncovers human-specific features of brain development
- Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19
- Single-cell multiomic profiling of human lungs reveals cell-type-specific and age-dynamic control of SARS-CoV2 host genes
December 01, 2024
New Projects (3)
- Cell type mapping reveals tissue niches and interactions in subcortical multiple sclerosis lesions
- Defining cellular complexity in human autosomal dominant polycystic kidney disease by multimodal single cell analysis
- Molecular Pathways of Colon Inflammation Induced by Cancer Immunotherapy
Updated projects (3):
- A blood atlas of COVID-19 defines hallmarks of disease severity and specificity
- A single cell atlas of human cornea that defines its development, limbal progenitor cells and their interactions with the immune cells.
- Asian Immune Diversity Atlas (AIDA): Single-cell analysis of human diversity in circulating immune cells (Japan cells)
October 31, 2024
New Raw Data, Metadata, and Contributor-generated Matrices (5)
- A temporal cortex cell atlas highlights gene expression dynamics during human brain maturation
- Evolution of neuronal cell classes and types in the vertebrate retina
- Single-Nucleus and In Situ RNA–Sequencing Reveal Cell Topographies in the Human Pancreas
- Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma
- Single-cell connectomic analysis of adult mammalian lungs
Updated Raw Data and Metadata
- Single-cell RNA sequencing of human kidney
- Single-cell sequencing unveils distinct immune microenvironments with CCR6-CCL20 crosstalk in human chronic pancreatitis
- Myofibroblast transcriptome indicates SFRP2hi fibroblast progenitors in systemic sclerosis skin
September 20, 2024
Release Highlights
- You can now request access to controlled project data and associated metadata.
Some genomic data available in the HCA portal may be regarded as sensitive/protected data under relevant privacy laws. These data are not available through the open-access tier, and instead can only be accessed via managed-access tier. This tier utilizes the Broad Institute’s Data Use Oversight System (DUOS). To learn more about working with such data, please check out this article on Requesting Access to Controlled Access Data.
New Raw Data, Metadata, and Contributor-generated Matrices (1)
September 13, 2024
New Raw Data, Metadata, and Contributor-generated Matrices (4)
- Activation of CD8+ T Cells in Chronic Obstructive Pulmonary Disease Lung
- An Atlas of Cells in the Human Tonsil
- Single-nucleus chromatin accessibility and transcriptomic map of breast tissues of women of diverse genetic ancestry
- Targeting the Immune-Fibrosis Axis in Myocardial Infarction and Heart Failure
August 5, 2024
Release Highlights
- New projects that are part of official HCA publications are now available:
New Raw Data, Metadata, and Contributor-generated Matrices (3)
- Dandelion uses the single-cell adaptive immune receptor repertoire to explore lymphocyte developmental origins
- Dynamic scRNA-seq of live human pancreatic slices reveals functional endocrine cell neogenesis through an intermediate ducto-acinar stage
- Human SARS-CoV-2 challenge resolves local and systemic response dynamics
Updated Raw Data and Metadata (30)
- A single-cell atlas of human teeth
- Single-Cell Transcriptomics of the Human Endocrine Pancreas
- Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis
- Single-cell multi-omics analysis of the immune response in COVID-19
- Single-cell transcriptomes identify human islet cell signatures and reveal cell-type–specific expression changes in type 2 diabetes
- Anomalous Epithelial Variations and Ectopic Inflammatory Response in Chronic Obstructive Pulmonary Disease
- Comparative Analysis and Refinement of Human PSC-Derived Kidney Organoid Differentiation with Single-Cell Transcriptomics
- Genetic ancestry effects on the response to viral infection are pervasive but cell type specific
- Human oral mucosa cell atlas reveals a stromal-neutrophil axis regulating tissue immunity
- Local and systemic responses to SARS-CoV-2 infection in children and adults
- Molecular characterization of foveal versus peripheral human retina by single-cell RNA sequencing
- Organoid single-cell genomic atlas uncovers human-specific features of brain development
- Pathogen-induced tissue-resident memory T H 17 (T RM 17) cells amplify autoimmune kidney disease
- Resolving the intertwining of inflammation and fibrosis in human heart failure at single-cell level
- Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations
- Single cell profiling of COVID-19 patients: an international data resource from multiple tissues
- Single-Cell Analysis of Human Pancreas Reveals Transcriptional Signatures of Aging and Somatic Mutation Patterns
- Single-Cell Heterogeneity Analysis and CRISPR Screen Identify Key β-Cell-Specific Disease Genes
- Single-Cell Transcriptome Profiling of Human Pancreatic Islets in Health and Type 2 Diabetes
- Single-Cell Transcriptomics of a Human Kidney Allograft Biopsy Specimen Defines a Diverse Inflammatory Response
- Single-cell RNA sequencing identifies celltype-specific cis-eQTLs and co-expression QTLs
- Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis
- Single-cell sequencing unveils distinct immune microenvironments with CCR6-CCL20 crosstalk in human chronic pancreatitis
- Single-cell transcriptomes of the human skin reveal age-related loss of fibroblast priming
- Single-cell transcriptomics reveals cell-type-specific diversification in human heart failure
- Single-nucleus cross-tissue molecular reference maps toward understanding disease gene function
- Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches
- The Tabula Sapiens: A multiple-organ, single-cell transcriptomic atlas of humans
- The single-cell transcriptomic landscape of early human diabetic nephropathy
- scRNA-seq assessment of the human lung, spleen, and esophagus tissue stability after cold preservation
June 28, 2024
Release Highlights
- A new project is available from the Oral & Craniofacial Biological Network.
New Raw Data, Metadata, and Contributor-generated Matrices (2)
- An Atlas of Cells in the Human Tonsil
- Single nucleus and spatial transcriptomic profiling of human healthy hamstring tendon
Updated Raw Data and Metadata (3)
- Recruited macrophages elicit atrial fibrillation
- Single-cell eQTL mapping identifies cell type-specific genetic control of autoimmune disease
- Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease
May 29, 2024
Release Highlights
- New projects are available from the Oral & Craniofacial and Pancreas Biological Networks.
New Raw Data, Metadata, and Contributor-generated Matrices (5)
- High-Resolution Transcriptomic Landscape of the Human Submandibular Gland
- Multimodal mapping of the tumor and peripheral blood immune landscape in human pancreatic cancer
- Single-cell multi-omics analysis of human pancreatic islets reveals novel cellular states in type 1 diabetes
- Single-nucleus RNA sequencing and spatial transcriptomics decode cell type-specific drivers underlying IBM pathogenesis
- Spatial transcriptomic profiling of human retinoblastoma
May 9, 2024
Release Highlights
- New projects are available from the Heart, Oral & Craniofacial, and Skin Biological Networks.
New Raw Data, Metadata, and Contributor-generated Matrices (9)
- Anatomically distinct fibroblast subsets determine skin autoimmune patterns
- Defining cardiac functional recovery in end-stage heart failure at single-cell resolution
- Distinct fibroblast progenitor subpopulation expedites regenerative mucosal healing by immunomodulation
- Salivary ZG16B expression loss follows exocrine gland dysfunction related to oral chronic graft-versus-host disease
- Single cell and lineage tracing studies reveal the impact of CD34+ cells on myocardial fibrosis during heart failure
- Single cell atlas of the human retina
- Single-cell multiomics of the human retina reveals hierarchical transcription factor collaboration in mediating cell type-specific effects of genetic variants on gene regulation
- Single-cell transcriptomic analysis of gingivo-buccal oral cancer reveals two dominant cellular programs
- Transcriptomic Mapping of Human Parotid Gland at Single-Cell Resolution
Updated Raw Data and Metadata (3)
- A cellular census of human lungs identifies novel cell states in health and in asthma
- A human cell atlas of the pressure-induced hypertrophic heart
- Cell-specific cis-regulatory elements and mechanisms of non-coding genetic disease in human retina and retinal organoids
April 8, 2024
Release Highlights
- New projects are available from the Eye, Heart, and Pancreas Biological Networks.
New Raw Data, Metadata, and Contributor-generated Matrices (4)
- A transcriptional cross species map of pancreatic islet cells
- Cardiac cell type-specific gene regulatory programs and disease risk association
- Recruited macrophages elicit atrial fibrillation
- Single-cell genomics improves the discovery of risk variants and genes of atrial fibrillation
Updated Raw Data and Metadata (4)
- A spatially resolved atlas of the human lung characterizes a gland-associated immune niche
- Integration of eQTL and a Single-Cell Atlas in the Human Eye Identifies Causal Genes for Age-Related Macular Degeneration
- Single cell profiling of human induced dendritic cells generated by direct reprogramming of embryonic fibroblasts
- Single-cell transcriptomic atlas of the human retina identifies cell types associated with age-related macular degeneration
March 20, 2024
Release Highlights
- New projects are available from the Eye, Heart, Lung, and Pancreas Biological Networks.
New Raw Data, Metadata, and Contributor-generated Matrices (11)
- Cell type-specific and disease-associated eQTL in the human lung
- A Single-Cell Transcriptome Atlas of the Human Pancreas
- Single-nucleus RNA sequencing of human pancreatic islets identifies novel gene sets and distinguishes β-cell subpopulations with dynamic transcriptome profiles
- Pseudotime Ordering of Single Human β-Cells Reveals States of Insulin Production and Unfolded Protein Response
- A human cell atlas of the pressure-induced hypertrophic heart
- Single-cell atlas of healthy human blood unveils age-related loss of NKG2C+GZMB-CD8+ memory T cells and accumulation of type 2 memory T cells
- Integrated multiomic characterization of congenital heart disease
- Single Nucleus RNA-sequencing Reveals Altered Intercellular Communication and Dendritic Cell Activation in Nonobstructive Hypertrophic Cardiomyopathy
- Single-nucleus RNA sequencing in ischemic cardiomyopathy reveals common transcriptional profile underlying end-stage heart failure
- Cell-specific cis-regulatory elements and mechanisms of non-coding genetic disease in human retina and retinal organoids
- Generation of human islet cell type-specific identity genesets
Updated Raw Data and Metadata (15)
- Spatial multi-omic map of human myocardial infarction
- Single cell transcriptional profiling of peripheral blood mononuclear cells (PBMCs) from mice flown on Rodent Research Reference Mission-2 (RRRM-2)
- GM-CSF-producing T helper cells: a distinct subset of T helper cells
- Single-cell transcriptomic and proteomic analysis of Parkinson’s disease brains
- Single-cell RNA-seq to decipher the subpopulations of human decidual leukocytes in normal and RSA pregnancies
- Retinal ganglion cell-specific genetic regulation in primary open angle glaucoma
- A single-cell transcriptome atlas of human early embryogenesis
- Single cell transcriptional profiling of spleens from mice flown on Rodent Research Reference Mission-2
- Spatial multiomics map of trophoblast development in early pregnancy
- Single-cell RNA sequencing of normal human kidney
- Cell Types of the Human Retina and Its Organoids at Single-Cell Resolution
- Single-cell multiome of the human retina and deep learning nominate causal variants in complex eye diseases
- Ischaemic sensitivity of human tissue by single cell RNA seq
- Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis
- Single-cell RNA-Seq Investigation of Foveal and Peripheral Expression in the Human Retina
January 16, 2023
Release Highlights
-
New projects are available from the Heart, Lung, Pancreas, and Skin Biological Networks.
-
A new managed access project, "Single-cell analysis reveals innate lymphoid cell lineage infidelity in atopic dermatitis", is available in the Data Portal.
New Raw Data, Metadata, and Contributor-generated Matrices (15)
- A Single-Cell Transcriptome Atlas of the Human Pancreas
- Identification of neural oscillations and epileptiform changes in human brain organoids
- Multi-scale spatial mapping of cell populations across anatomical sites in healthy human skin and basal cell carcinoma
- Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis
- Single Cell Multiome Atlas of the Human Fetal Retina
- Single-cell RNA sequencing reveals markers of disease progression in primary cutaneous T-cell lymphoma
- Single-cell analysis reveals innate lymphoid cell lineage infidelity in atopic dermatitis
- Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19
- Single-cell transcriptomics combined with interstitial fluid proteomics defines cell type-specific immune regulation in atopic dermatitis
- Single-cell transcriptomics provides insights into hypertrophic cardiomyopathy
- Single-cell transcriptomics reveals distinct effector profiles of infiltrating T cells in lupus skin and kidney
- Single-nucleus profiling of human dilated and hypertrophic cardiomyopathy
- Spatially resolved multiomics of human cardiac niches
- The serine proteases dipeptidyl-peptidase 4 and urokinase are key molecules in human and mouse scar formation
- Transcriptional and Cellular Diversity of the Human Heart
Updated Raw Data and Metadata (26)
- Ischaemic sensitivity of human tissue by single cell RNA seq
- A Single-Cell Atlas of the Human Healthy Airways
- A cellular census of human lungs identifies novel cell states in health and in asthma
- A single-cell transcriptome atlas of the adult human retina
- A spatially resolved atlas of the human lung characterizes a gland-associated immune niche
- Airspace Macrophages and Monocytes Exist in Transcriptionally Distinct Subsets in Healthy Adults
- An Optimized Tissue Dissociation Protocol for Single-Cell RNA Sequencing Analysis of Fresh and Cultured Human Skin Biopsies
- Cryopreservation and post-thaw characterization of dissociated human islet cells
- Dissecting the cellular specificity of smoking effects and reconstructing lineages in the human airway epithelium
- Human prefrontal cortex gene regulatory dynamics from gestation to adulthood at single-cell resolution
- Identification of a unique subset of tissue-resident memory CD4+ T cells in Crohn’s disease
- Integrative analysis of cell state changes in lung fibrosis with peripheral protein biomarkers
- Pro-inflammatory T helper 17 directly harms oligodendrocytes in neuroinflammation
- SARS‐CoV‐2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells
- Single Cell RNA-seq reveals ectopic and aberrant lung resident cell populations in Idiopathic Pulmonary Fibrosis
- Single-Cell Transcriptome and Downstream Analyses of Human Stomach Reveal the Cell Diversity and Developmental Features of Gastric and Metaplastic Mucosae
- Single-Cell Transcriptomic Analysis Identifies a Unique Pulmonary Lymphangioleiomyomatosis Cell
- Single-Cell Transcriptomic Analysis of Human Lung Provides Insights into the Pathobiology of Pulmonary Fibrosis
- Single-cell RNA-sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis
- Single-cell Transcriptome Atlas of the Human Corpus Cavernosum
- Single-cell and single-nucleus RNA-seq uncovers shared and distinct axes of variation in dorsal LGN neurons in mice, non-human primates, and humans
- Single-cell transcriptomics reveals unique features of human pancreatic islet cell subtypes
- Spatio-temporal immune zonation of the human kidney
- The Immune Atlas of Human Deciduas With Unexplained Recurrent Pregnancy Loss
- The local and systemic response to SARS-CoV-2 infection in children and adults
- Transcriptional analysis of cystic fibrosis airways at single-cell resolution reveals altered epithelial cell states and composition
December 4, 2023
Release Highlights
- A redesigned HCA Data Portal is live, featuring the first atlases from the Lung and Nervous System Biological Networks. The new portal contains a page for each HCA Biological Network, as well as a new guide, Exploring Biological Network and Atlas Data.
New Raw Data, Metadata, and Contributor-generated Matrices (8)
- A spatially resolved single-cell genomic atlas of the adult human breast
- An Optimized Tissue Dissociation Protocol for Single-Cell RNA Sequencing Analysis of Fresh and Cultured Human Skin Biopsies
- Classification of human chronic inflammatory skin disease based on single-cell immune profiling
- Crosstalk of astrocyte, microglia, and neuronal subtypes correlates to Alzheimer’s disease pathology
- Neuronal defects in a human cellular model of 22q11.2 deletion syndrome
- Single cell transcriptional zonation of human psoriasis skin identifies an alternative immunoregulatory axis conducted by skin resident cells
- Transcriptomic analysis of the ocular posterior segment completes a cell atlas of the human eye
- αβγδ T cells play a vital role in fetal human skin development and immunity
Updated Raw Data and Metadata (4)
- Cell atlas of the human ocular anterior segment: Tissue-specific and shared cell types
- Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19
- Harnessing Expressed Single Nucleotide Variation and Single Cell RNA Sequencing To Define Immune Cell Chimerism in the Rejecting Kidney Transplant
- Single-cell RNA sequencing reveals the effects of chemotherapy on human pancreatic adenocarcinoma and its tumor microenvironment
October 31, 2023
Release Highlights
-
A new project that is part of an official HCA publication is now available: "Single nucleus transcriptome and chromatin accessibility of postmortem human pituitaries reveal diverse stem cell regulatory mechanisms"
-
New projects are available from the Brain Biological Network.
New Raw Data, Metadata, and Contributor-generated Matrices (7)
- A single-cell atlas of entorhinal cortex from individuals with Alzheimer's disease reveals cell-type-specific gene expression regulation
- Dysregulation of brain and choroid plexus cell types in severe COVID-19
- Molecular and cellular evolution of the primate dorsolateral prefrontal cortex
- Single nucleus transcriptome and chromatin accessibility of postmortem human pituitaries reveal diverse stem cell regulatory mechanisms
- Single-Cell Transcriptome and Downstream Analyses of Human Stomach Reveal the Cell Diversity and Developmental Features of Gastric and Metaplastic Mucosae
- Single-cell and single-nucleus RNA-seq uncovers shared and distinct axes of variation in dorsal LGN neurons in mice, non-human primates, and humans
- Single-cell genomic profiling of human dopamine neurons identifies a population that selectively degenerates in Parkinson's disease
Updated Raw Data and Metadata (5)
- A Cellular Anatomy of the Normal Adult Human Prostate and Prostatic Urethra
- Joint profiling of chromatin accessibility and gene expression in thousands of single cells
- Re-evaluation of human BDCA-2+ DC during acute sterile skin inflammation
- Single-cell transcriptome profiling of an adult human cell atlas of 15 major organs
- Single-cell transcriptomics uncovers distinct molecular signatures of stem cells in chronic myeloid leukemia
September 27, 2023
Release Highlights
- New projects are available from the Brain and Lung Biological Networks.
New Raw Data, Metadata, and Contributor-generated Matrices (8)
- A molecular cell atlas of the human lung from single-cell RNA sequencing
- Allergic inflammatory memory in human respiratory epithelial progenitor cells
- Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain
- Molecular characterization of selectively vulnerable neurons in Alzheimer's disease
- Phenotype molding of stromal cells in the lung tumor microenvironment
- Single-cell RNA sequencing reveals the effects of chemotherapy on human pancreatic adenocarcinoma and its tumor microenvironment
- Single-nucleus transcriptome analysis of human brain immune response in patients with severe COVID-19
- The ZIP8/SIRT1 axis regulates alveolar progenitor cell renewal in aging and idiopathic pulmonary fibrosis
Updated Raw Data and Metadata (5)
- Reconstitution of a functional human thymus by postnatal stromal progenitor cells and natural whole-organ scaffolds
- The Single Cell Transcriptomic Landscape of Early Human Diabetic Nephropathy
- A multi-omic single-cell landscape of human gynecologic malignancies
- Discriminating mild from critical COVID-19 by innate and adaptive immune single-cell profiling of bronchoalveolar lavages
- Harnessing Expressed Single Nucleotide Variation and Single Cell RNA Sequencing To Define Immune Cell Chimerism in the Rejecting Kidney Transplant
August 28, 2023
New Raw Data, Metadata, and Contributor-generated Matrices (5)
- Cellular development and evolution of the mammalian cerebellum
- Dysregulated lung stroma drives emphysema exacerbation by potentiating resident lymphocytes to suppress an epithelial stem cell reservoir
- Mapping single-cell transcriptomes in the intra-tumoral and associated territories of kidney cancer
- Single-cell atlas of bronchoalveolar lavage from preschool cystic fibrosis reveals new cell phenotypes
- Single-cell sequencing of human midbrain reveals glial activation and a Parkinson-specific neuronal state
Updated Raw Data and Metadata (23)
- A Cellular Atlas of Pitx2-Dependent Cardiac Development
- A Single-Cell Transcriptomic Map of the Human and Mouse Pancreas Reveals Inter- and Intra-cell Population Structure
- A human single cell atlas of the substantia nigra reveals novel cell specific pathways associated with the genetic risk of Parkinson’s disease and neuropsychiatric disorders
- A single-cell survey of the human first-trimester placenta and decidua
- CD27hiCD38hi plasmablasts are activated B cells of mixed origin with distinct function
- Colonic single epithelial cell census reveals goblet cell drivers of barrier breakdown in inflammatory bowel disease
- Defining human mesenchymal and epithelial heterogeneity in response to oral inflammatory disease
- Defining the Activated Fibroblast Population in Lung Fibrosis Using Single Cell Sequencing
- Dissecting the clonal nature of allelic expression in somatic cells by single-cell RNA-seq
- Dissecting the protein and transcriptional responses of human immune cells to T cell and monocyte specific activation
- GM-CSF-producing T helper cells: a distinct subset of T helper cells
- Massively parallel single-cell RNA-seq analysis of 26,677 pancreatic islets cells from both healthy and type II diabetic (T2D) donors
- Single cell RNA-sequencing of human tonsil Innate lymphoid cells (ILCs)
- Single cell analysis of human fetal liver captures the transcriptional profile of hepatobiliary hybrid progenitors
- Single-cell RNA sequencing reveals the heterogeneity of liver-resident immune cells in human
- Single-cell Transcriptome Analysis Reveals Dynamic Cell Populations and Differential Gene Expression Patterns in Control and Aneurysmal Human Aortic Tissue
- Single-cell transcriptomes of the aging human skin reveal loss of fibroblast priming
- Single-nucleus cross-tissue molecular reference maps to decipher disease gene function
- Transcriptomic and clonal characterization of T cells in the human central nervous system
- A single-cell reference map of transcriptional states for human blood and tissue T cell activation
- A single-cell regulatory map of postnatal lung alveologenesis in humans and mice
- Single-cell analysis of human non-small cell lung cancer lesions refines tumor classification and patient stratification
- Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors
July 27, 2023
New Raw Data, Metadata, and Contributor-generated Matrices (21)
- Single-cell multiomic profiling of human lungs reveals cell-type-specific and age-dynamic control of SARS-CoV2 host genes
- Single-cell genomics improves the discovery of risk variants and genes of cardiac traits
- Dissecting the treatment-naive ecosystem of human melanoma brain metastasis
- Collagen-producing lung cell atlas identifies multiple subsets with distinct localization and relevance to fibrosis
- Village in a dish: a model system for population-scale hiPSC studies
- Single-Cell Transcriptomics Reveal Immune Mechanisms of the Onset and Progression of IgA Nephropathy
- The immune cell landscape in kidneys of patients with lupus nephritis
- Multimodal single cell sequencing of human diabetic kidney disease implicates chromatin accessibility and genetic background in disease progression
- Single-cell analysis reveals fibroblast heterogeneity and myofibroblasts in systemic sclerosis-associated interstitial lung disease
- Lung transplantation for patients with severe COVID-19
- COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas
- Single-Cell Transcriptomic Analysis Identifies a Unique Pulmonary Lymphangioleiomyomatosis Cell
- Marked regional glial heterogeneity in the human white matter of the central nervous system
- Discriminating mild from critical COVID-19 by innate and adaptive immune single-cell profiling of bronchoalveolar lavages
- A single cell atlas of the human liver tumor microenvironment
- Single-cell analysis of human non-small cell lung cancer lesions refines tumor classification and patient stratification
- Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia
- Regenerative lineages and immune-mediated pruning in lung cancer metastasis
- Single cell landscape of mesenchymal and endothelial cells in healthy human liver
- Differential cell composition and split epidermal differentiation in human palm, sole, and hip skin
- Human distal lung maps and lineage hierarchies reveal a bipotent progenitor
Updated Raw Data and Metadata (4)
- Cell atlas of the human ocular anterior segment: Tissue-specific and shared cell types
- COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets
- A multi-omics atlas of the human retina at single-cell resolution
- AIDA
June 26, 2023
Release Highlights
-
New projects are available from the Blood, Eye, Gut, Kidney, Lung, and Skin Biological Networks.
-
A new managed access project, "A multi-omic single-cell landscape of human gynecologic malignancies", is available in the Data Portal.
-
A project previously removed from the Data Portal has been re-imported and is available for download:
New Raw Data, Metadata, and Contributor-generated Matrices (14)
- A multi-omic single-cell landscape of human gynecologic malignancies
- A multi-omics atlas of the human retina at single-cell resolution
- A single-cell regulatory map of postnatal lung alveologenesis in humans and mice
- Airspace Macrophages and Monocytes Exist in Transcriptionally Distinct Subsets in Healthy Adults
- Characterization of altered molecular mechanisms in Parkinson's disease through cell type-resolved multiomics analyses
- Dynamic changes in human single cell transcriptional signatures during fatal sepsis
- Human melanocyte development and melanoma dedifferentiation at single-cell resolution
- Immune signatures underlying post-acute COVID-19 lung sequelae
- Impaired local intrinsic immunity to SARS-CoV-2 infection in severe COVID-19
- Single cell transcriptomic analysis of the immune cell compartment in the human small intestine and in Celiac disease
- Single cell transcriptomics identifies focal segmental glomerulosclerosis remission endothelial biomarker
- Single-cell analysis of the cellular heterogeneity and interactions in the injured mouse spinal cord
- Spatially resolved human kidney multi-omics single cell atlas highlights the key role of the fibrotic microenvironment in kidney disease progression
- The landscape of immune dysregulation in Crohn's disease revealed through single-cell transcriptomic profiling in the ileum and colon
Updated Raw Data and Metadata (2)
- A single cell atlas of the human proximal epididymis reveals cell-type specific functions and distinct roles for CFTR
- Time-resolved systems immunology reveals a late juncture linked to fatal COVID-19
June 13, 2023
Release Highlights
-
New projects are available from the Eye, Gut, and Kidney Biological Networks.
-
Several projects were removed from the Data Portal while metadata is being updated. They will be added to the portal again during an upcoming release.
-
COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets (UUID: 61515820-5bb8-45d0-8d12-f0850222ecf0)
-
Single-cell analysis of the cellular heterogeneity and interactions in the injured mouse spinal cord (UUID: e6773550-c1a6-4949-8643-1a3154cf2670)
-
New Raw Data, Metadata, and Contributor-generated Matrices (16)
- Benchmarking Single-Cell RNA Sequencing Protocols for Cell Atlas Projects
- Cell Atlas of The Human Fovea and Peripheral Retina
- Construction of A Human Cell Landscape by Single-cell mRNA-seq
- Cross-tissue immune cell analysis reveals tissue-specific features in humans
- Differential pre-malignant programs and microenvironment chart distinct paths to malignancy in human colorectal polyps
- Identification of a unique subset of tissue-resident memory CD4+ T cells in Crohn’s disease
- Integration of eQTL and a Single-Cell Atlas in the Human Eye Identifies Causal Genes for Age-Related Macular Degeneration
- Intrarenal Single-Cell Sequencing of Hepatitis B Virus Associated Membranous Nephropathy
- Pathogen-induced tissue-resident memory TH17 (TRM17) cells amplify autoimmune kidney disease
- Single cell RNA sequencing to dissect the molecular heterogeneity in lupus nephritis
- Single-cell analyses of Crohn's disease tissues reveal intestinal intraepithelial T cells heterogeneity and altered subset distributions
- Single-cell analyses of renal cell cancers reveal insights into tumor microenvironment, cell of origin, and therapy response
- Single-cell multiome of the human retina and deep learning nominate causal variants in complex eye diseases
- Single-cell profiling of healthy human kidney reveals features of sex-based transcriptional programs and tissue-specific immunity
- Single-cell sequencing reveals novel cellular heterogeneity in uterine leiomyomas
- Single-cell transcriptomic atlas of the human retina identifies cell types associated with age-related macular degeneration
Updated Raw Data and Metadata (4)
- Single cell sequencing identifies novel sub-populations of breast cancer cells selected under hypoxia
- Single-cell RNA sequencing identifies cell type-specific cis-eQTLs and co-expression QTLs
- A single-cell transcriptomic atlas of the human ciliary body
- Integrated analysis of multimodal single-cell data
May 11, 2023
Release Highlights
- A new project, "Mucosal Profiling of Pediatric-Onset Colitis and IBD Reveals Common Pathogenics and Therapeutic Pathways", is available from the Gut Biological Network.
New Raw Data, Metadata, and Contributor-generated Matrices (3)
- A Partial Picture of the Single-Cell Transcriptomics of Human IgA Nephropathy
- Mucosal Profiling of Pediatric-Onset Colitis and IBD Reveals Common Pathogenics and Therapeutic Pathways
- Single-Cell Atlas of Lineage States, Tumor Microenvironment, and Subtype-Specific Expression Programs in Gastric Cancer
Updated Raw Data and Metadata (2)
- Severe COVID-19 Is Marked by a Dysregulated Myeloid Cell Compartment
- Vento-tormo: Single cell profiling of COVID-19 patients: an international data resource from multiple tissues
March 31, 2023
Release Highlights
-
A full project metadata spreadsheet is now available for several projects in the Data Portal. You can download the spreadsheet from the Metadata page for any project where it is available by selecting the download icon under Metadata.
-
A new managed access project, "Spatially organized multicellular immune hubs in human colorectal cancer", is available in the Data Portal.
-
New projects are available from the Blood, Gut, and Kidney Biological Networks.
New Raw Data, Metadata, and Contributor-generated Matrices (10)
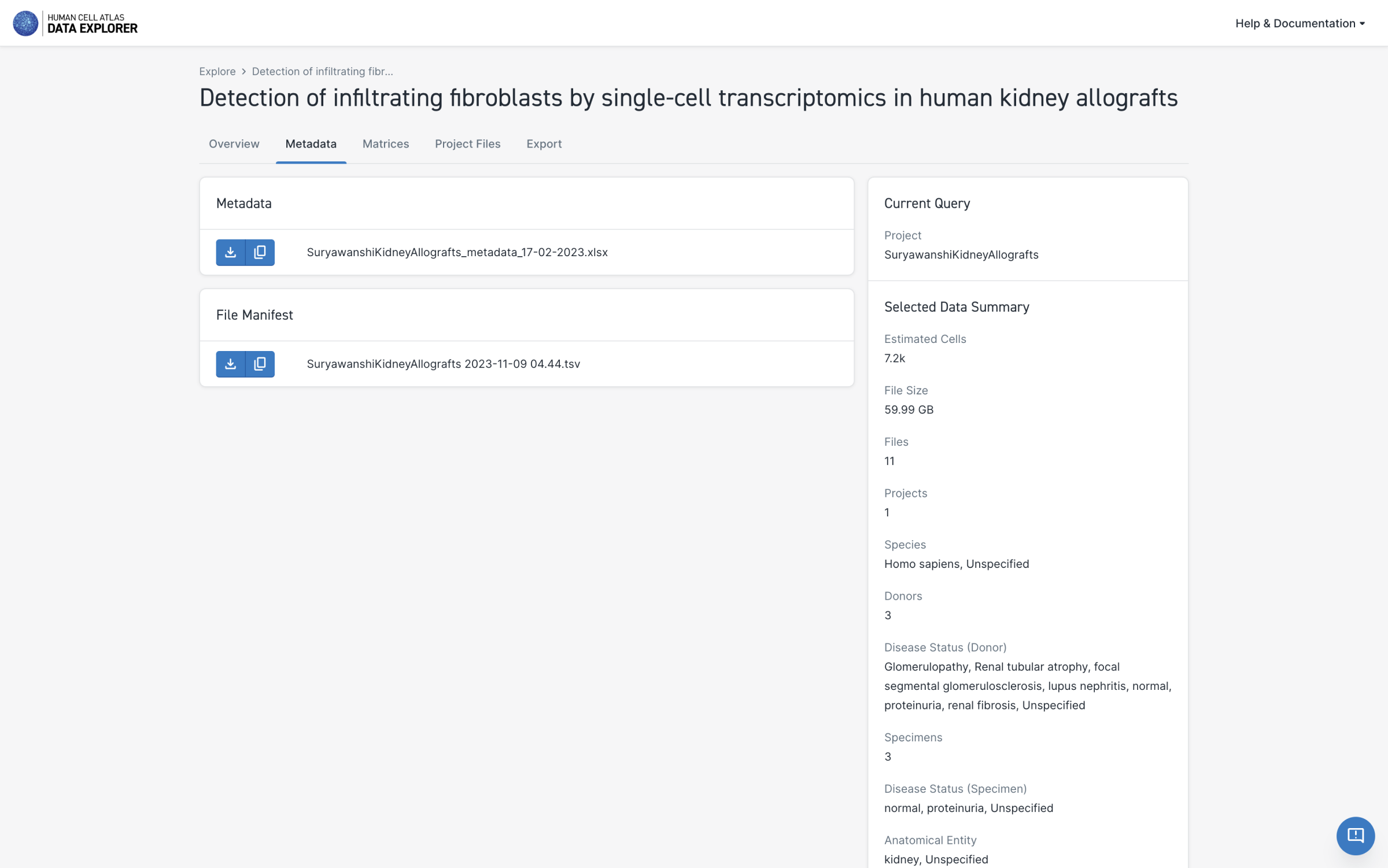
- Detection of infiltrating fibroblasts by single-cell transcriptomics in human kidney allografts
- Harnessing Expressed Single Nucleotide Variation and Single Cell RNA Sequencing To Define Immune Cell Chimerism in the Rejecting Kidney Transplant
- Heterogeneity and clonal relationships of adaptive immune cells in ulcerative colitis revealed by single-cell analyses
- Single-Cell RNA Sequencing in Multiple Pathologic Types of Renal Cell Carcinoma Revealed Novel Potential Tumor-Specific Markers
- Single-cell multiomics revealed the dynamics of antigen presentation, immune response and T cell activation in the COVID-19 positive and recovered individuals
- Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors
- Spatially organized multicellular immune hubs in human colorectal cancer
- Time-resolved systems immunology reveals a late juncture linked to fatal COVID-19
- Transcriptional changes in the mammary gland during lactation revealed by single cell sequencing of cells from human milk
- Ulcerative colitis is characterized by a plasmablast-skewed humoral response associated with disease activity
February 24, 2023
Release Highlights
-
Thanks to feedback from members of the HCA community, we’ve added two new metadata fields, Genome version and Patch version, to projects in the Data Portal! Check out the field descriptions in the Metadata Dictionary to learn more or start exploring HCA projects in the Data Portal!
-
New data is available from researchers in Australia; see the project "Human prefrontal cortex gene regulatory dynamics from gestation to adulthood at single-cell resolution."
-
A new project that is part of an official HCA publication is now available: "Comprehensive cell atlas of the first-trimester developing human brain."
-
New projects are available from the Blood and Kidney Biological Networks.
New Raw Data, Metadata, and Contributor-generated Matrices (11)
- A blood atlas of COVID-19 defines hallmarks of disease severity and specificity
- Comprehensive cell atlas of the first-trimester developing human brain
- Human prefrontal cortex gene regulatory dynamics from gestation to adulthood at single-cell resolution
- Integrated analysis of multimodal single-cell data
- Severe COVID-19 Is Marked by a Dysregulated Myeloid Cell Compartment
- Single-cell Transcriptome Atlas of Testis Aging
- Single-cell eQTL mapping identifies cell type-specific genetic control of autoimmune disease
- Single-cell protein activity analysis identifies recurrence-associated renal tumor macrophages
- Single-cell sequencing links multiregional immune landscapes and tissue-resident T cells in ccRCC to tumor topology and therapy efficacy
- The single cell atlas of human endometrial stromal cells
- Vento-tormo: Single cell profiling of COVID-19 patients: an international data resource from multiple tissues
Updated Raw Data and Metadata (3)
- A spatially resolved atlas of the human lung characterizes a gland-associated immune niche
- Plasticity of distal nephron epithelia from human kidney organoids enables the induction of ureteric tip and stalk
- Comparison of manual and two types of bioprinted kidney organoids by single cell RNA-seq
December 20, 2022
Release Highlights
- Contributors have added new donors, data, and metadata to what was previously the prototype data set "Census of Immune cells"; the updated project, "A single cell immune cell atlas of human hematopoietic system," now has over 1,453,784 analyzed cells.
- New data is available from researchers in China; see the project "Immune cell profiling of preeclamptic pregnant and postpartum women by single-cell RNA sequencing."
Raw Data, Metadata, and Contributor-generated Matrices (10)
- A Single-Cell Transcriptome Atlas of Glia Diversity in the Human Hippocampus across the Lifespan and in Alzheimer’s Disease
- A single-cell transcriptomic atlas of the human ciliary body
- HCA seed network precise tumor-nephrectomy samples
- Immune cell profiling of preeclamptic pregnant and postpartum women by single-cell RNA sequencing
- Massively Parallel Single Nucleus Transcriptional Profiling Defines Spinal Cord Neurons and Their Activity during Behavior
- Single cell transcriptomics reveals spatial and temporal dynamics of gene expression in the developing mouse spinal cord
- Single-cell analysis of the cellular heterogeneity and interactions in the injured mouse spinal cord
- Single-cell dissection of the human brain vasculature
- Single-nuclei profiling of aged skeletal muscle
- Single-nucleus transcriptomic atlas of spinal cord neuron in human
Updated Raw Data and Metadata (6)
- A single cell immune cell atlas of human hematopoietic system
- Fate-mapping within human iPSC-derived kidney organoids reveals conserved mammalian nephron progenitor lineage relationships
- Kidney micro-organoids in suspension culture as a scalable source of human pluripotent stem cell-derived kidney cells
- RNA-Seq from human innate lymphoid cells (ILCs) transitional populations
- Genome-Wide DNA Hypermethylation in the Wound-Edge of Chronic Wound Patients Opposes Closure by Impairing Epithelial to Mesenchymal Transition
- Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy
November 28, 2022
Release Highlights
- New single-cell transcriptomic data is available for adult mouse spinal cord; see the project Single-cell transcriptomic analysis of the adult mouse spinal cord reveals molecular diversity of autonomic and skeletal motor neurons.
- Single-cell data derived from peritoneal and ovarian lesions of participants with endometriosis is available in the project Single cell analysis of endometriosis reveals a coordinated transcriptional program driving immunotolerance and angiogenesis across eutopic and ectopic tissues.
New Raw Data, Metadata, and Contributor-generated Matrices (10)
- Cell atlas of the human ocular anterior segment: Tissue-specific and shared cell types
- Dissecting the cellular specificity of smoking effects and reconstructing lineages in the human airway epithelium
- SARS-CoV-2 infection of the oral cavity and saliva
- Single cell RNA-seq analysis of adult and paediatric IDH-wildtype Glioblastomas
- Single cell analysis of endometriosis reveals a coordinated transcriptional program driving immunotolerance and angiogenesis across eutopic and ectopic tissues
- Single-Cell Lymphocyte Heterogeneity in Advanced Cutaneous T-cell Lymphoma Skin Tumors
- Single-cell atlas of diverse immune populations in the advanced biliary tract cancer microenvironment
- Single-cell transcriptomic analyses provide insights into the developmental origins of neuroblastoma
- Single-cell transcriptomic analysis of the adult mouse spinal cord reveals molecular diversity of autonomic and skeletal motor neurons
- The regulatory landscapes of human ovarian ageing
Updated Metadata (3)
- A spatial multi-omics atlas of the human lung reveals a novel immune cell survival niche
- Cellular heterogeneity of human fallopian tubes in normal and hydrosalpinx disease states identified by scRNA-seq
- A single-cell reference map of transcriptional states for human blood and tissue T cell activation
October 19, 2022
Release Highlights
-
New COVID-19 data is available in the project COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets.
-
The new project Genome-Wide DNA Hypermethylation in the Wound-Edge of Chronic Wound Patients Opposes Closure by Impairing Epithelial to Mesenchymal Transition combines spatial transcriptomic data with methylome data.
New Raw Data, Metadata, and Contributor-generated Matrices (4)
- COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets
- Differential dynamics of response at single cell resolution following CAR-T therapy in refractory B-cell lymphoma
- Genome-Wide DNA Hypermethylation in the Wound-Edge of Chronic Wound Patients Opposes Closure by Impairing Epithelial to Mesenchymal Transition
- Single-cell analysis of childhood leukemia reveals a link between developmental states and ribosomal protein expression as a source of intra-individual heterogeneity
Updated Metadata (2)
- Expansion of Fcγ Receptor IIIa-Positive Macrophages, Ficolin 1-Positive Monocyte-Derived Dendritic Cells, and Plasmacytoid Dendritic Cells Associated With Severe Skin Disease in Systemic Sclerosis
- A single-cell and single-nucleus RNA-seq toolbox for fresh and frozen human tumors
September 26, 2022
Release Highlights
- The Data Portal now hosts over 30 million cells! Thank you to the HCA community and all of the contributors that helped us reach this milestone!
New Raw Data, Metadata, and Contributor-generated Matrices (12)
- A Single-Cell Characterization of Human Post-implantation Embryos Cultured In Vitro Delineates Morphogenesis in Primary Syncytialization
- A Single-Cell Transcriptomic Atlas of Human Skin Aging
- A human breast atlas integrating single-cell proteomics and transcriptomics
- Defining Transcriptional Signatures of Human Hair Follicle Cell States
- Expansion of Fcγ Receptor IIIa-Positive Macrophages, Ficolin 1-Positive Monocyte-Derived Dendritic Cells, and Plasmacytoid Dendritic Cells Associated With Severe Skin Disease in Systemic Sclerosis
- Mapping the developing human immune system across organs
- Robust temporal map of human in vitro myelopoiesis using single-cell genomics
- Single cell transcriptomics of human epidermis identifies basal stem cell transition states
- Single nucleus RNA-sequencing defines unexpected diversity of cholinergic neuron types in the adult mouse spinal cord
- Single-Cell RNA Sequencing Maps Endothelial Metabolic Plasticity in Pathological Angiogenesis
- Single-cell transcriptomic and proteomic analysis of Parkinson’s disease brains
- The myogenesis program drives clonal selection and drug resistance in rhabdomyosarcoma
Updated Metadata (5)
- Single-cell RNA-Seq reveals a developmental atlas of human prefrontal cortex
- Profiling of transcribed cis-regulatory elements in single cells
- Single-Cell Transcriptomic Analysis of Human Lung Provides Insights into the Pathobiology of Pulmonary Fibrosis
- Spatial multi-omic map of human myocardial infarction
- Spatial proteogenomics reveals distinct and evolutionarily-conserved hepatic macrophage niches
August 23, 2022
Release Highlights
-
There are two new managed access projects available in the Browser:
-
A new COVID-19 dataset has been added to the HCA Data Portal as part of the project Longitudinal Multi-omics Analyses Identify Responses of Megakaryocytes, Erythroid Cells, and Plasmablasts as Hallmarks of Severe COVID-19.
-
New raw data derived from populations in Asia and Australia are now available.
New Raw Data, Metadata, and Contributor-generated Matrices (9)
- CD90 marks a mesenchymal program in human thymic epithelial cells in vitro and in vivo
- Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts
- Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets
- Longitudinal Multi-omics Analyses Identify Responses of Megakaryocytes, Erythroid Cells, and Plasmablasts as Hallmarks of Severe COVID-19
- Reconstitution of a functional human thymus by postnatal stromal progenitor cells and natural whole-organ scaffolds
- Refining Colorectal Cancer Classification and Clinical Stratification Through a Single-Cell Atlas
- Single Cell Transcriptomics Reveals Cell Type Specific Diversification in Human Heart Failure
- Single-cell Transcriptome Atlas of the Human Corpus Cavernosum
- The immune cell atlas of human neuroblastoma
Updated Metadata (39)
- A Human Liver Cell Atlas reveals Heterogeneity and Epithelial Progenitors
- A Single-Cell Transcriptomic Map of the Human and Mouse Pancreas Reveals Inter- and Intra-cell Population Structure
- A cellular census of human lungs identifies novel cell states in health and in asthma
- A single cell atlas of human cornea that defines its development, limbal progenitor cells and their interactions with the immune cells
- A single-cell and single-nucleus RNA-seq toolbox for fresh and frozen human tumors
- A spatial multi-omics atlas of the human lung reveals a novel immune cell survival niche
- Assessing the relevance of organoids to model inter-individual variation
- Blood and immune development in human fetal bone marrow and Down syndrome
- Construction of a single-cell transcriptomic atlas of 58,243 liver cells from 4 donors and 4 recipient liver transplasnt patients to investigate early allograft dysfunction (EAD)
- Decoding Human Megakaryocyte Development
- Developmental cell programs are co-opted in inflammatory skin disease
- Human cerebral organoids recapitulate gene expression programs of fetal neocortex development
- Improving fibroblast characterization using single-cell RNA sequencing: an optimized tissue disaggregation and data processing pipeline
- Integrated single cell analysis of blood and cerebrospinal fluid leukocytes in multiple sclerosis
- Integrative analysis of cell state changes in lung fibrosis with peripheral protein biomarkers
- Integrative computational analysis of the substantia nigra pars compacta and the locus coeruleus
- Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex
- Modelling the impact of decidual senescence on embryo implantation in human endometrial assembloids
- Molecular Architecture of the Mouse Nervous System
- Molecular and functional heterogeneity of IL-10-producing CD4+ T cells
- Population-scale single-cell RNA-seq profiling across dopaminergic neuron differentiation
- Profiling of transcribed cis-regulatory elements in single cells
- SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues
- Sampling time-dependent artifacts in single-cell genomics studies
- Single Cell RNA-seq reveals ectopic and aberrant lung resident cell populations in Idiopathic Pulmonary Fibrosis
- Single Cell RNAseq of primary pulmonary endothelial cells
- Single-Cell Transcriptomic Analysis of Human Lung Provides Insights into the Pathobiology of Pulmonary Fibrosis
- Single-Cell Transcriptomics Uncovers Zonation of Function in the Mesenchyme during Liver Fibrosis
- Single-cell RNA sequencing identifies cell type-specific cis-eQTLs and co-expression QTLs
- Single-cell RNA-Seq of human hepatocyte-derived liver progenitor-like cells
- Single-cell RNA-sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis
- Single-cell analysis of human B cell maturation predicts how antibody class switching shapes selection dynamics
- Single-cell transcriptomes of the aging human skin reveal loss of fibroblast priming
- Single-nucleus RNA-seq in the post-mortem brain in major depressive disorder
- Spatial and single-cell transcriptional profiling identifies functionally distinct human dermal fibroblast subpopulations
- Spatial multi-omic map of human myocardial infarction
- Spatial proteogenomics reveals distinct and evolutionarily-conserved hepatic macrophage niches
- The Human and Mouse Enteric Nervous System at Single-Cell Resolution
- Transcriptional analysis of cystic fibrosis airways at single-cell resolution reveals altered epithelial cell states and composition
July 19, 2022
Release Highlights
-
New combined 10x Visium and single-cell RNA sequencing data derived from human ureter tissue are available in the project Ureter single-cell and spatial mapping reveal cell types, architecture, and signaling networks.
-
Multiple new projects feature data related to SARS-CoV-2 infection:
New Raw Data, Metadata, and Contributor-generated Matrices (6)
- Lineage recording in human cerebral organoids
- Single Cell, Single Nucleus and Spatial RNA Sequencing of the Human Liver Identifies Hepatic Stellate Cell and Cholangiocyte Heterogeneity
- Single-cell RNA-seq reveals cell type-specific molecular and genetic associations to lupus
- Single-cell meta-analysis of SARS-CoV-2 entry genes across tissues and demographics
- The local and systemic response to SARS-CoV-2 infection in children and adults
- Ureter single-cell and spatial mapping reveal cell types, architecture, and signaling networks
Updated Metadata (4)
- A single-cell and single-nucleus RNA-seq toolbox for fresh and frozen human tumors
- Defining the Activated Fibroblast Population in Lung Fibrosis Using Single Cell Sequencing
- Single-cell RNA Sequencing of human microglia from post mortem Alzheimers Disease CNS tissue
- Spatial multi-omic map of human myocardial infarction
June 29, 2022
Release Highlights
-
New data is available from research groups in Japan, China, and South Korea:
- scRNA-seq experiments on human foreskin fibroblasts, foreskin fibroblasts induced directly to retina pigment epithelium, and induced pluripotent stem cell derived retina pigment epithelium
- Single-cell RNA-seq of bone marrow cells from aplastic anemia patient and healthy donor
- Memory-like natural killer cells facilitate effector functions of daratumumab in multiple myeloma
-
There are now seven 10x Visium spatial transcriptomic projects available in the Data Browser. See the most recent one: A human fetal lung cell atlas uncovers proximal-distal gradients of differentiation and key regulators of epithelial fates.
New Raw Data, Metadata, and Contributor-generated Matrices (11)
- A human fetal lung cell atlas uncovers proximal-distal gradients of differentiation and key regulators of epithelial fates
- A single-cell and single-nucleus RNA-seq toolbox for fresh and frozen human tumors
- Decoding Human Megakaryocyte Development
- Memory-like natural killer cells facilitate effector functions of daratumumab in multiple myeloma
- Profiling of transcribed cis-regulatory elements in single cells
- Single-cell RNA-seq of bone marrow cells from aplastic anemia patient and healthy donor
- Single-cell RNA-seq to decipher the subpopulations of human decidual leukocytes in normal and RSA pregnancies
- Single-cell deciphering of human fetal innate lymphoid cell development
- Single-cell sequencing unveils the heterogeneity of nonimmune cells in chronic apical periodontitis
- The Human and Mouse Enteric Nervous System at Single-Cell Resolution
- scRNA-seq experiments on human foreskin fibroblasts, foreskin fibroblasts induced directly to retina pigment epithelium, and induced pluripotent stem cell derived retina pigment epithelium
Updated Metadata (28)
- Assessing the relevance of organoids to model inter-individual variation
- A single cell atlas of human teeth
- Blood and immune development in human fetal bone marrow and Down syndrome
- Capturing human trophoblast development with naive pluripotent stem cells in vitro
- Dissecting the Global Dynamic Molecular Profiles of Human Fetal Kidney Development by Single-Cell RNA Sequencing
- Extreme heterogeneity of influenza virus infection in single cells
- Human photoreceptor cells from different macular subregions have distinct transcriptional profiles
- Integrative computational analysis of the substantia nigra pars compacta and the locus coeruleus
- Population-scale single-cell RNA-seq profiling across dopaminergic neuron differentiation
- Single Cell RNA Sequencing of Human Milk-Derived Cells Reveals Sub-Populations of Mammary Epithelial Cells with Molecular Signatures of Progenitor and Mature States: a Novel, Non-invasive Framework for Investigating Human Lactation Physiology
- Single Cell RNA-Seq profiling human embryonic kidney cortex cells
- Single cell RNA-seq of HeLa CCL2
- Single cell RNAseq of human nail unit defines RSPO4 onychofibroblasts and SPINK6 nail epithelium
- Single cell sequencing identifies novel sub-populations of breast cancer cells selected under hypoxia
- Single cell transcriptional profiling of peripheral blood mononuclear cells (PBMCs) from mice flown on Rodent Research Reference Mission-2 (RRRM-2)
- Single cell transcriptional profiling of spleens from mice flown on Rodent Research Reference Mission-2
- Single-Cell RNA Sequencing Reveals Human iPS Cell-derived Alveolar Type 1 Cells in Alveolar Organoids
- Single-Cell Transcriptomic Profiling of Human Dental Pulp in Sound and Carious Teeth: A Pilot Study
- Single-cell Transcriptome Analysis Reveals an Anomalous Epithelial Variation and Ectopic Inflammatory Response in Chronic Obstructive Pulmonary Disease
- Single-cell transcriptome profiling of an adult human cell atlas of 15 major organs
- Single-cell transcriptomics of normal pancreas, intraductal papillary mucinous neoplasm, and pancreatic adenosquamous carcinoma reveals the heterogeneous progression of pancreatic ductal and stromal cells
- Single-cell transcriptomics of the human retinal pigment epithelium and choroid in health and macular degeneration
- Single-cell transcriptomics reveals unique features of human pancreatic islet cell subtypes
- Spatial multi-omic map of human myocardial infarction
- Spatial proteogenomics reveals distinct and evolutionarily-conserved hepatic macrophage niches
- Spatially distinct reprogramming of the tumor microenvironment based on tumor invasion in diffuse-type gastric cancers
- The Immune Atlas of Human Deciduas With Unexplained Recurrent Pregnancy Loss
- The effect of Mtb aggregation on monocyte-derived macrophage transcription profiles
June 9, 2022
Release Highlights
-
Contributions of spatial transcriptomic data (like 10x Visium) continue to climb; for example, see the project Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex.
-
New raw data and contributor matrices shed light on disease processes; see the project Single-Cell Transcriptomics of Human Substantia Nigra in Parkinson's Disease for a cellular atlas of the human substantia nigra in health and Parkinson’s Disease.
-
Multi-modal data provides a more complete understanding of cellular function. The new project Spatial proteogenomics reveals distinct and evolutionarily-conserved hepatic macrophage niches combines the power of Visium and CITE-seq.
New Raw Data, Metadata, and Contributor-generated Matrices (13)
- A Human Liver Cell Atlas reveals Heterogeneity and Epithelial Progenitors
- A single-cell atlas of breast cancer cell lines to study tumour heterogeneity and drug response
- CITE-seq of in vitro control and memory-like human NK cells
- Combinatorial transcription factor profiles predict mature and functional human islet α and β cells
- RNA-Seq from human innate lymphoid cells (ILCs) transitional populations
- SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues
- Single Cell RNA-seq reveals ectopic and aberrant lung resident cell populations in Idiopathic Pulmonary Fibrosis
- Single-Cell Transcriptomics of Human Substantia Nigra in Parkinson's Disease
- Single-cell analysis of gastric pre-cancerous and cancer lesions reveals cell lineage diversity and intratumoral heterogeneity
- Skin single cell universe identifies IL-1B/IL23A co-producing CD14+ type 3 dendritic cells in psoriasis
- Spatial multi-omic map of human myocardial infarction
- Spatial proteogenomics reveals distinct and evolutionarily-conserved hepatic macrophage niches
- Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex
Updated Raw Data, Metadata, Contributor-generated Matrices (4)
- Single-cell analysis of human B cell maturation predicts how antibody class switching shapes selection dynamics
- Improving fibroblast characterization using single-cell RNA sequencing: an optimized tissue disaggregation and data processing pipeline
- Single-cell RNA Sequencing of human microglia from post mortem Alzheimers Disease CNS tissue
- Single-cell profiling of human subventricular zone progenitors identifies SFRP1 as a target for stimulating progenitor activation
April 20, 2022
Release Highlights
-
Multiple projects now include DOI information in the metadata and project page:
-
New raw imaging data is available for the projects:
-
Two new research projects include data from mice that traveled to the International Space Station:
-
A new project that is part of an official HCA publication is now available: A proximal-to-distal survey of healthy adult human small intestine and colon epithelium by single-cell transcriptomics
New Raw Data, Metadata, and Contributor-generated Matrices (19):
- A proximal-to-distal survey of healthy adult human small intestine and colon epithelium by single-cell transcriptomics
- A single-cell transcriptome atlas of human early embryogenesis
- A spatial multi-omics atlas of the human lung reveals a novel immune cell survival niche
- Cells of the human intestinal tract mapped across space and time
- Construction of a single-cell transcriptomic atlas of 58,243 liver cells from 4 donors and 4 recipient liver transplasnt patients to investigate early allograft dysfunction (EAD)
- Improving fibroblast characterization using single-cell RNA sequencing: an optimized tissue disaggregation and data processing pipeline
- Integrated single-cell transcriptomics and epigenomics reveals strong germinal center–associated etiology of autoimmune risk loci
- Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex
- Molecular signatures of inflammatory profile and B-cell function in SFTS patients
- Retinal ganglion cell-specific genetic regulation in primary open angle glaucoma
- Single cell RNA-seq of HeLa CCL2
- Single cell transcriptional profiling of peripheral blood mononuclear cells (PBMCs) from mice flown on Rodent Research Reference Mission-2 (RRRM-2)
- Single cell transcriptional profiling of spleens from mice flown on Rodent Research Reference Mission-2
- Single cell transcriptome atlases of the developing mouse and human spinal cord
- Single-cell RNA sequencing identifies cell type-specific cis-eQTLs and co-expression QTLs
- Single-cell RNA-Seq reveals a developmental atlas of human prefrontal cortex
- Single-nucleus RNA-seq in the post-mortem brain in major depressive disorder
- Single-nucleus transcriptomic profiles of glial cells in human dorsal root ganglion and spinal cord
- Spatiotemporal Analysis of Human Intestinal Development at Single Cell Resolution - scRNA-Seq
Updated Raw Data, Metadata, Contributor-generated Matrices (10):
- Blood and immune development in human fetal bone marrow and Down syndrome
- Nuclei multiplexing with barcoded antibodies for single-nucleus genomics
- A cell atlas of human thymic development defines T cell repertoire formation
- A comprehensive single cell transcriptional landscape of human hematopoietic progenitors
- A single cell atlas of human cornea that defines its development, limbal progenitor cells and their interactions with the immune cells
- Decoding human fetal liver haematopoiesis
- Mapping the temporal and spatial dynamics of the human endometrium in vivo and in vitro
- Modelling the impact of decidual senescence on embryo implantation in human endometrial assembloids
- Single-Cell Transcriptomic Profiling of Human Dental Pulp in Sound and Carious Teeth: A Pilot Study
- Single-cell analysis reveals the continuum of human lympho-myeloid progenitor cells
March 30, 2022
Release Highlights
-
A new managed access project, A cellular census of human lungs identifies novel cell states in health and in asthma, is available in the Browser.
-
A new COVID-19 dataset has been added to the HCA Data Portal as part of the project Single-cell RNA sequencing of urinary cells reveals distinct cellular diversity in COVID-19-associated AKI.
-
New raw data derived from populations in Africa, Asia, and Europe are now available.
New Raw Data, Metadata, and Contributor-generated Matrices (11):
- A cellular census of human lungs identifies novel cell states in health and in asthma
- A single cell atlas of human cornea that defines its development, limbal progenitor cells and their interactions with the immune cells
- Conserved cell types with divergent features in human versus mouse cortex
- Modelling the impact of decidual senescence on embryo implantation in human endometrial assembloids
- Molecular Architecture of the Mouse Nervous System
- Single-cell RNA sequencing of urinary cells reveals distinct cellular diversity in COVID-19-associated AKI
- Single-cell RNA-sequencing reveals pervasive but highly cell type-specific genetic ancestry effects on the response to viral infection
- Single-cell analysis of human B cell maturation predicts how antibody class switching shapes selection dynamics
- Single-cell profiling of human subventricular zone progenitors identifies SFRP1 as a target for stimulating progenitor activation
- Spatially distinct reprogramming of the tumor microenvironment based on tumor invasion in diffuse-type gastric cancers
- Microglia Require CD4 T Cells to Complete the Fetal-to-Adult Transition
Updated Raw Data, Metadata, Contributor-generated Matrices (6):
- Assessing the relevance of organoids to model inter-individual variation
- Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19
- Integrative computational analysis of the substantia nigra pars compacta and the locus coeruleus
- Single-cell RNA-Seq of human hepatocyte-derived liver progenitor-like cells
- The Developmental Heterogeneity of Human Natural Killer Cells Defined by Single-cell Transcriptome
- The cellular immune response to COVID-19 deciphered by single cell multi-omics across three UK centres
February 14, 2022
Release Highlights
-
We've created a new slack channel for the HCA Data Portal community! Join the channel, say hello, and follow for the latest announcements, updates, and opportunities to connect with fellow HCA Data Portal users.
-
The Data Portal header has been updated to include new features:
-
A global search bar that allows users to search across Projects, Guides and Metadata Schema.
-
Direct links to the HCA Twitter, GitHub repository, and slack channel.

-
-
There are three new managed access projects available in the Browser:
-
The first dataset of iPSC-derived tenocytes has been added to the HCA Data Portal as part of the project Single cell RNAseq analysis of the developmental trajectory of iPSC-derived tenocytes.
-
A new dataset generated using Fluidigm C1 sequencing has been added to the HCA Data Portal as part of the project Pseudo-temporal ordering of individual cells reveals regulators of differentiation.
-
New data from across the globe has been added to the HCA Data Portal, including datasets from Japan, China, and Thailand.
New Raw Data, Metadata, and Contributor-generated Matrices (14):
- A multi-omics atlas of the human retina at single-cell resolution
- Cell Types of the Human Retina and Its Organoids at Single-Cell Resolution
- Dissecting transcriptional and chromatin accessibility heterogeneity of proliferating cone precursors in human retinoblastoma tumours by single cell sequencing
- Integrated scRNA-Seq Identifies Human Postnatal Thymus Seeding Progenitors and Regulatory Dynamics of Differentiating Immature Thymocytes
- Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis
- Pseudo-temporal ordering of individual cells reveals regulators of differentiation
- Single cell RNA sequencing of human failing heart
- Single cell RNAseq analysis of the developmental trajectory of iPSC-derived tenocytes
- Single-Cell Transcriptomic Profiling of Human Dental Pulp in Sound and Carious Teeth: A Pilot Study
- Single-Cell Transcriptomics Uncovers Zonation of Function in the Mesenchyme during Liver Fibrosis
- Single-cell transcriptome analysis reveals the dynamics of human immune cells during early fetal skin development
- Single-nucleus RNA-seq profiling of the human primary motor cortex in amyotrophic lateral sclerosis and frontotemporal lobar degeneration
- Spatial and single-cell transcriptional landscape of human cerebellar development
- The Tabula Sapiens: a single cell transcriptomic atlas of multiple organs from individual human donors
New HCA Data Portal Analysis (19):
- Cryopreservation and post-thaw characterization of dissociated human islet cells
- Dissecting the clonal nature of allelic expression in somatic cells by single-cell RNA-seq
- Melanoma infiltration of stromal and immune cells
- Re-evaluation of human BDCA-2+ DC during acute sterile skin inflammation
- Single cell RNA-seq of human pancreatic endocrine cells from Juvenile, adult control and type 2 diabetic donors
- Single cell RNA-sequencing of human tonsil Innate lymphoid cells (ILCs)
- Single cell analysis of human fetal liver captures the transcriptional profile of hepatobiliary hybrid progenitors
- Single cell sequencing identifies novel sub-populations of breast cancer cells selected under hypoxia
- Single-Cell RNAseq analysis of diffuse neoplastic infiltrating cells at the migrating front of human glioblastoma
- Single-Cell Transcriptomics Reveals a Population of Dormant Neural Stem Cells that Become Activated upon Brain Injury
- Single-cell RNA-seq reveals heterogeneity within human pre-cDCs
- Single-cell analysis reveals the continuum of human lympho-myeloid progenitor cells
- Single-cell omics reveal human mononuclear phagocyte heterogeneity and inflammatory DC in health and disease
- Single-cell transcriptomics reveals unique features of human pancreatic islet cell subtypes
- Single-cell transcriptomics uncovers distinct molecular signatures of stem cells in chronic myeloid leukemia
- Spatial and single-cell transcriptional profiling identifies functionally distinct human dermal fibroblast subpopulations
- Systematic comparative analysis of single cell RNA-sequencing methods
- Tracing pluripotency of human early embryos and embryonic stem cells by single cell RNA-seq
- Transcriptomes of 1,529 individual cells from 88 human preimplantation embryos
December 20, 2021
Release Highlights
-
The Data Portal header has been updated with a new link to the Updates page so that viewers can readily follow release notes and other community announcements.
-
There are two new managed access projects available in the Browser:
-
Three projects have new uniform matrices generated with the updated Optimus pipeline that includes STARsolo software:
- Single-cell transcriptomics uncovers human corneal limbal stem cells and their differentiation trajectory.
- A single-cell atlas of the healthy breast tissues reveals clinically relevant clusters of breast epithelial cells.
- Single cell RNA-Seq of E18.5 developing mouse kidney and human kidney organoids.
-
Several new projects also include data generated with multiple sequencing modalities (i.e. CITE-seq, 10x 3', and scATAC):
New Raw Data, Metadata, and Contributor-generated Matrices (21):
- A Single-Cell Atlas of the Human Healthy Airways
- A comprehensive single cell transcriptional landscape of human hematopoietic progenitors
- A single-cell atlas of chromatin accessibility in the human genome
- An organoid and multi-organ developmental cell atlas reveals multilineage fate specification in the human intestine
- Blood and immune cell development in human fetal bone marrow and in Down syndrome
- CD27hiCD38hi plasmablasts are activated B cells of mixed origin with distinct function
- Co-evolution of tumor and immune cells during progression of multiple myeloma
- Extreme heterogeneity of influenza virus infection in single cells
- Hypertension delays viral clearance and exacerbates airway hyperinflammation in patients with COVID-19
- Identification of distinct synovial fibroblast subsets associated with pain and progression of knee osteoarthritis using single-cell RNA sequencing
- Integrative analysis of cell state changes in lung fibrosis with peripheral protein biomarkers
- Kidney organoid reproducibility across multiple human iPSC lines and diminished off target cells after transplantation revealed by single cell transcriptomics
- Modeling Hepatoblastoma: Identification of Distinct Tumor Cell Populations and Key Genetic Mechanisms through Single Cell Sequencing (scRNA-seq)
- SARS‐CoV‐2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells
- Sampling time-dependent artifacts in single-cell genomics studies
- Single-Cell Transcriptomic Analysis of Human Lung Provides Insights into the Pathobiology of Pulmonary Fibrosis
- Single-cell ATAC sequencing reveals the mechanism of human breast cancer metastasis
- Single-cell RNA sequencing of human femoral head in vivo
- Single-cell RNA sequencing of peripheral blood reveals immune cell signatures in Alzheimer's disease
- Single-cell sequencing unveils distinct immune microenvironment in human chronic pancreatitis
- Using single-nucleus RNA-sequencing to interrogate transcriptomic profiles of archived human pancreatic islets
Updated Metadata, Raw Data, Contributor-generated Matrices, or HCA Data Portal Analysis (7):
- A single-cell atlas of the healthy breast tissues reveals clinically relevant clusters of breast epithelial cells
- AIDA pilot data
- Distinct microbial and immune niches of the human colon
- Single cell RNA-Seq of E18.5 developing mouse kidney and human kidney organoids
- Single-cell RNA-seq analysis throughout a 125-day differentiation protocol that converted H1 human embryonic stem cells to a variety of ventrally-derived cell types
- Single-cell transcriptomics uncovers human corneal limbal stem cells and their differentiation trajectory
- Stress-induced RNA–chromatin interactions promote endothelial dysfunction
November 11, 2021
Release Highlights
- The HCA Data Portal team has reanalyzed the first of the HCA Data Portal 1.0 Smart-seq2 projects (Single cell RNA sequencing of multiple myeloma II). Cell-by-gene matrices are available on the project page and were produced with the latest Smart-seq Multi-Sample pipeline for uniform analysis.
- Contributor-generated matrices are available for the project Single-nucleus cross-tissue molecular reference maps to decipher disease gene function which is associated with a Human Cell Atlas preprint publication.
- New raw data has been added for the project Defining human mesenchymal and epithelial heterogeneity in response to oral inflammatory disease. as part of the HCA Oral & Craniofacial Biological Network.
- The first onychofibroblasts dataset has been added to the HCA Data Portal as part of the project Single cell RNAseq of human nail unit defines RSPO4 onychofibroblasts and SPINK6 nail epithelium.
- New patch-seq data is available in the project Cryopreservation and post-thaw characterization of dissociated human islet cells.
- A content description for contributor- and HCA Data Portal-generated matrices is now available on each Data Portal project's matrix download page.
- The submitted and last updated dates are added to the projects list and Data Portal Explore page. We will make this sortable by this field in a future release.
- New and updated projects for the latest HCA Data Portal Platform release are now indicated next to the project name on
the Explore
page (as shown below).
New Raw Data, Metadata, and Contributor-generated Matrices (14)
- Cellular heterogeneity of human fallopian tubes in normal and hydrosalpinx disease states identified by scRNA-seq
- Cryopreservation and post-thaw characterization of dissociated human islet cells
- Defining human mesenchymal and epithelial heterogeneity in response to oral inflammatory disease
- Differentiation of Human Intestinal Organoids with Endogenous Vascular Endothelial Cells
- Healthy human kidney cell type single cell RNA-seq data
- Human photoreceptor cells from different macular subregions have distinct transcriptional profiles
- In Vitro and In Vivo Development of the Human Airway at Single-Cell Resolution
- Mapping Development of the Human Intestinal Niche at Single-Cell Resolution
- Mapping the temporal and spatial dynamics of the human endometrium in vivo and in vitro
- Pro-inflammatory T helper 17 directly harms oligodendrocytes in neuroinflammation
- Single Cell RNAseq of primary pulmonary endothelial cells
- Single cell RNAseq of human nail unit defines RSPO4 onychofibroblasts and SPINK6 nail epithelium
- Single cell sequencing identifies novel sub-populations of breast cancer cells selected under hypoxia
- Single-nucleus cross-tissue molecular reference maps to decipher disease gene function
Updated Metadata, Raw Data, Contributor-generated Matrices, or HCA Data Portal Analysis (2)
October 13, 2021
Release Highlights
-
Project downloads are now available from the project page. Download the files directly to your local machine via curl or export them to the cloud-based bioinformatics platform Terra by selecting the download next to the new "Terra Workspace" icon.
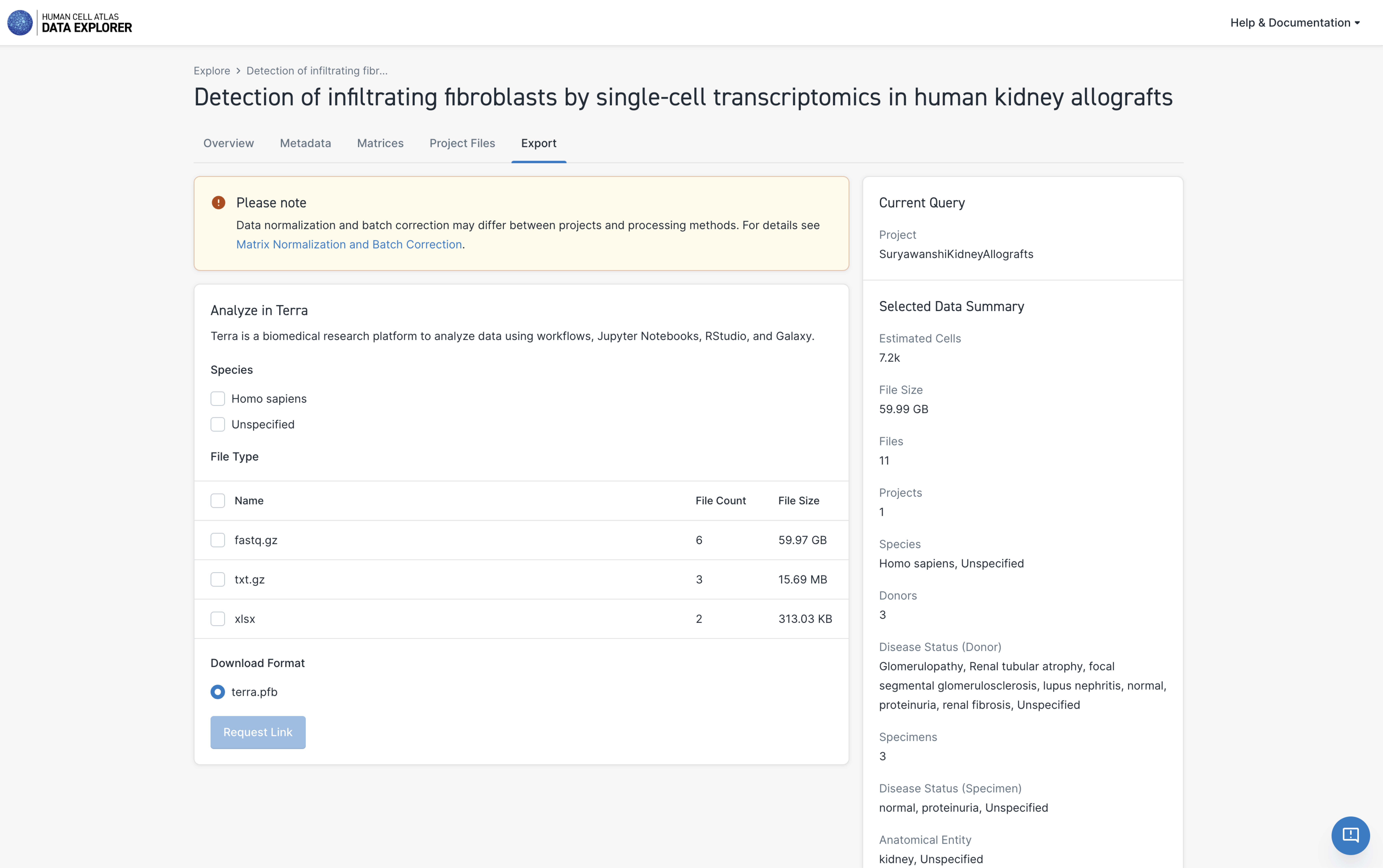
-
The projects A human cell atlas of fetal gene expression, Single-cell atlas of early human brain development highlights heterogeneity of human neuroepithelial cells and early radial glia, and COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis contain new managed-access data.
-
The Data Portal team has corrected linking issues associated with the project A single cell atlas of human teeth.
-
A new HumanCellAtlas slack channel (#dcp-help) is available to help you troubleshoot Data Portal questions with both the Data Portal team and community and give you the latest updates on Data Portal events and data.
New Raw Data, Metadata, and Contributor-generated Matrices (14)
- A Single-cell Transcriptomic Atlas of Human Intervertebral Disc
- A human cell atlas of fetal gene expression
- COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis
- Kidney micro-organoids in suspension culture as a scalable source of human pluripotent stem cell-derived kidney cells
- Lifespan charactarization of the nasal mucosa
- Population-scale single-cell RNA-seq profiling across dopaminergic neuron differentiation
- Single cell RNA-seq of brain glioblastoma samples
- Single cell RNA-sequencing on healthy and IPF lung mesenchymal cells
- Single cell atlas of the healthy breast
- Single-cell RNA-Seq of human hepatocyte-derived liver progenitor-like cells
- Single-cell analysis of Crohn's disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy
- Single-cell atlas of early human brain development highlights heterogeneity of human neuroepithelial cells and early radial glia
- Single-cell atlas of human oral mucosa reveals a stromal-neutrophil axis in tissue immunity regulation
- Single-cell transcriptomics of normal pancreas, intraductal papillary mucinous neoplasm, and pancreatic adenosquamous carcinoma reveals the heterogeneous progression of pancreatic ductal and stromal cells
New HCA Data Portal Analysis (2)
- Census of Immune Cells
- Integrative computational analysis of the substantia nigra pars compacta and the locus coeruleus
Updated Metadata, Contributor-generated Matrices, or HCA Data Portal Analysis (20)
- A single cell atlas of human teeth
- Capturing human trophoblast development with naive pluripotent stem cells in vitro
- Dissecting the Global Dynamic Molecular Profiles of Human Fetal Kidney Development by Single-Cell RNA Sequencing
- Female human primordial germ cells display X-chromosome dosage compensation despite the absence of X-inactivation [Female]
- Female human primordial germ cells display X-chromosome dosage compensation despite the absence of X-inactivation [Male]
- Resolving cellular and molecular diversity along the hippocampal anterior-to-posterior axis in humans
- Single Cell RNA Sequencing of Human Milk-Derived Cells Reveals Sub-Populations of Mammary Epithelial Cells with Molecular Signatures of Progenitor and Mature States: a Novel, Non-invasive Framework for Investigating Human Lactation Physiology
- Single Cell RNA-Seq profiling human embryonic kidney cortex cells
- Single Cell RNA-Seq profiling of human embryonic kidney outer and inner cortical cells and kidney organoid cells
- Single-Cell RNA Sequencing Reveals Human iPS Cell-derived Alveolar Type 1 Cells in Alveolar Organoids
- Single-Cell RNA-Seq Analysis of cells from human urine
- Single-cell RNA sequencing of human nasal swab from healthy donors
- Single-cell RNA-sequencing reveals the landscape of cervical cancer heterogeneity
- Single-cell Transcriptome Analysis Reveals an Anomalous Epithelial Variation and Ectopic Inflammatory Response in Chronic Obstructive Pulmonary Disease
- Single-cell reconstruction of follicular remodeling in the human adult ovary
- Single-cell transcriptomics of the human retinal pigment epithelium and choroid in health and macular degeneration
- Single-cell transcriptomics reveals unique features of human pancreatic islet cell subtypes
- The Developmental Heterogeneity of Human Natural Killer Cells Defined by Single-cell Transcriptome
- The Immune Atlas of Human Deciduas With Unexplained Recurrent Pregnancy Loss
- The effect of Mtb aggregation on monocyte-derived macrophage transcription profiles
September 17, 2021
New Raw Data
The HCA Data Portal has added raw data for the following 14 new projects:
- A molecular single-cell lung atlas of lethal COVID-19
- Capturing human trophoblast development with naive pluripotent stem cells in vitro
- Comprehensive single-cell transcriptome analysis reveals heterogeneity in endometrioid adenocarcinoma tissues
- Fate-mapping within human iPSC-derived kidney organoids reveals conserved mammalian nephron progenitor lineage relationships
- Female human primordial germ cells display X-chromosome dosage compensation despite the absence of X-inactivation [Female]
- Female human primordial germ cells display X-chromosome dosage compensation despite the absence of X-inactivation [Male]
- Plasticity of distal nephron epithelia from human kidney organoids enables the induction of ureteric tip and stalk
- Resolving cellular and molecular diversity along the hippocampal anterior-to-posterior axis in humans
- Single Cell RNA Sequencing of Human Milk-Derived Cells Reveals Sub-Populations of Mammary Epithelial Cells with Molecular Signatures of Progenitor and Mature States: a Novel, Non-invasive Framework for Investigating Human Lactation Physiology
- Single Cell RNA-Seq profiling of human embryonic kidney outer and inner cortical cells and kidney organoid cells
- Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment
- Single-Cell RNA-Seq Analysis of cells from human urine
- Single-cell Transcriptome Analysis Reveals an Anomalous Epithelial Variation and Ectopic Inflammatory Response in Chronic Obstructive Pulmonary Disease
- The effect of Mtb aggregation on monocyte-derived macrophage transcription profiles
Updated Data
The following 3 projects have updated files:
- A single-cell atlas of the healthy breast tissues reveals clinically relevant clusters of breast epithelial cells
- Single cell RNA-Seq of E18.5 developing mouse kidney and human kidney organoids
- Single-cell transcriptomics uncovers human corneal limbal stem cells and their differentiation trajectory
HCA Data Portal now contains data for 14 million estimated cells
August 25, 2021
New Raw Data
The HCA Data Portal has added raw data for the following 12 new projects:
- AIDA pilot data
- Comparison of manual and two types of bioprinted kidney organoids by single cell RNA-seq
- Developmental cell programs are co-opted in inflammatory skin disease
- Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer
- Single-Cell RNA Sequencing Reveals Human iPS Cell-derived Alveolar Type 1 Cells in Alveolar Organoids
- Single-cell RNA sequencing of human nasal swab from healthy donors
- Single-cell transcriptome profiling of an adult human cell atlas of 15 major organs
- Single-cell transcriptome profiling of the vaginal wall in women with severe anterior vaginal prolapse
- Single-cell transcriptomics of East-Asian pancreatic islets cells
- The Developmental Heterogeneity of Human Natural Killer Cells Defined by Single-cell Transcriptome
- The Immune Atlas of Human Deciduas With Unexplained Recurrent Pregnancy Loss
- Transcriptional analysis of cystic fibrosis airways at single-cell resolution reveals altered epithelial cell states and composition
New Contributor Data
The following projects have new contributor-generated matrix files:
- Integrated single cell analysis of blood and cerebrospinal fluid leukocytes in multiple sclerosis
- Single-cell transcriptomes of the aging human skin reveal loss of fibroblast priming
Browser Updates
Selecting "Normal" Samples
To enable easier discovery of normal tissues, we've modified the Specimen Disease search facet on the Explore page so that the "Normal" option is now at the top of the drop-down.
Selecting Files by "Content Description"
The File facet has a new Content Description column to enable searching for files by the type of data they contain.
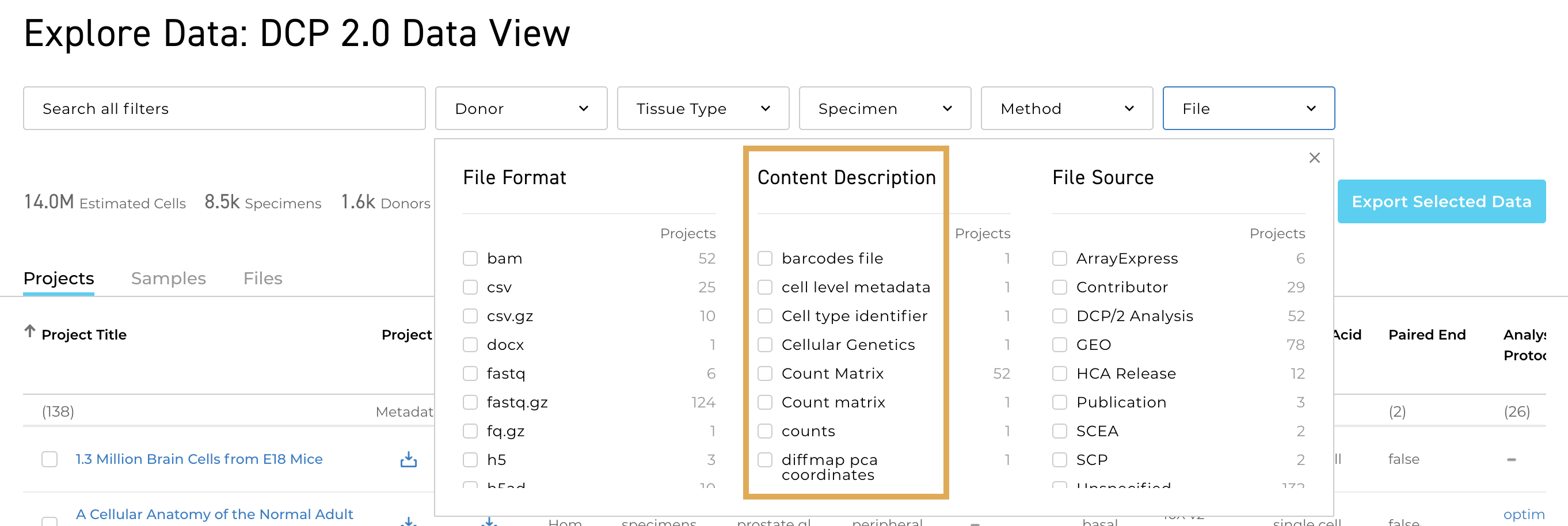
New Managed Access and Seed Network projects
July 23, 2021
The HCA Data Portal has added its first managed access project.
Contributor-generated cell-by-gene count matrices and associated non-personally identifiable metadata are now available to the whole HCA community from within the HCA Data Portal.
Additionally, the HCA Data Portal now has the first project data from the HCA Seed Networks, which are further developing the Human Cell Atlas by combining the expertise of experimental scientists, software engineers, and physicians.
Managed Access Projects
Managed access raw data is available from the Synapse database upon successful completion of a data use certificate. To find supplementary links to the managed access datasets in Synapse, navigate to the project detail page in the Data Portal.
HCA Seed Networks Projects
- Single cell transcriptional and chromatin accessibility profiling redefine cellular heterogeneity in the adult human kidney
- Stress-induced RNA–chromatin interactions promote endothelial dysfunction
- A single-cell atlas of the healthy breast tissues reveals clinically relevant clusters of breast epithelial cells
Additional New or Updated Projects
Projects with new raw data and/or contributor-generated matrices:
- Integrated scRNA-Seq Identifies Human Postnatal Thymus Seeding Progenitors and Regulatory Dynamics of Differentiating Immature Thymocytes
- Single-cell transcriptomic atlas of the human endometrium during the menstrual cycle
- Single-cell reconstruction of follicular remodeling in the human adult ovary
- Single Cell RNA-Seq profiling human embryonic kidney cortex cells
- Dissecting the Global Dynamic Molecular Profiles of Human Fetal Kidney Development by Single-Cell RNA Sequencing
- Establishing Cerebral Organoids as Models of Human-Specific Brain Evolution
- Single-cell transcriptomics of the human retinal pigment epithelium and choroid in health and macular degeneration
- A single cell atlas of human teeth
- Single cell RNA-Seq of E18.5 developing mouse kidney and human kidney organoids
- Single-cell RNA sequencing of normal human kidney
- Single-cell RNA-sequencing reveals the landscape of cervical cancer heterogeneity
- Single-cell transcriptomics reveals unique features of human pancreatic islet cell subtypes
- Re-evaluation of human BDCA-2+ DC during acute sterile skin inflammation
- Molecular and functional heterogeneity of IL-10-producing CD4+ T cells
- Single-cell transcriptomes of the aging human skin reveal loss of fibroblast priming
Projects with new uniformly processed data:
- Single-cell analysis reveals congruence between kidney organoids and human fetal kidney
- A Cellular Anatomy of the Normal Adult Human Prostate and Prostatic Urethra
- The emergent landscape of the mouse gut endoderm at single-cell resolution
- Microglia Require CD4 T Cells to Complete the Fetal-to-Adult Transition
- High throughput error corrected Nanopore single cell transcriptome sequencing
- Colonic single epithelial cell census reveals goblet cell drivers of barrier breakdown in inflammatory bowel disease
Data Portal now contains data for 13.5 million cells
June 16, 2021
- Raw data and contributor-generated matrices are available for 15 new projects. These single-cell data are derived from:
- Single-cell and single-nucleus
- 10x 3' v2 and v3 3' and 10x 5' chemistry, Fluidigm C1, Smart-seq2, and sci-CAR ATAC sequencing technologies
- Adipose tissue, blood, bone marrow, CSF, heart, kidney, lung, oral cavity, ovary, pancreas, prostate, skeletal muscle, testis, and umbilical vein
- Disease states including COVID-19 infection, intracranial hypertension, multiple sclerosis, and renal cell carcinoma
- Developmental stages including fetal, child, adolescent, and adult
- Standardized data, including aligned BAMs and cell-by-gene count matrices (Loom format), are available for 8 additional projects with data derived from:
- 10x v2 and v3 3' sequencing technologies
- Brain (superior parietal cortex, middle temporal gyrus, and temporal cortex), epididymis, immune tissue, cortex of the kidney organoid, lymph nodes, placenta (chorionic villus and decidua), spine, testis, and thymus
- Disease states including Alzheimer disease and cognitive impairment with or without cerebellar ataxia
- New Jupyter Notebook tutorials for analyzing standardized HCA Data Portal matrix files are available in an Intro-to-HCA-data-on-Terra workspace on the cloud-based platform Terra.
- After registering, you can try the step-by-step instructions for importing HCA data and analyzing in common community tools such as Bioconductor, Cumulus, Pegasus, Scanpy, and Seurat.
Raw sequencing data and contributor-generated matrices for 23 new projects available for download
May 10, 2021
- Data Portal now has data for 12.2 million cells, including new standardized analyses for 13 projects as well as raw sequencing data and contributor-generated matrices for 23 new projects.
- The standardized data, including aligned BAMs and cell-by-gene count matrices (Loom format), are derived from:
- Human and mouse
- Single-cell and single-nucleus
- 10x V2 and V3 3' chemistry
- Blood, brain (including substantia nigra, developing hippocampus, cortex, retina), liver, lung, mouth, skeletal muscle, skin, spleen, and developing thymus
- Disease states including HIV, drug hypersensitivity syndrome, multiple sclerosis, and thoracic aortic aneurysm
- The 23 new projects and contributor-generated matrices include data derived from:
- Human and mouse
- Smart-seq2, 10x V2 and V3 3' chemistry, Drop-seq, and Fluidigm C1 sequencing methods
- Blood, bone marrow, brain, cord blood, epididymis, fetal gonads, immune organ, kidney organoid, lymph nodes, pancreas, skin, and trachea
- Disease states including Alzheimer's Disease, multiple sclerosis, autoimmune encephalitis, and type 2 diabetes
- The Matrix Overview guide has been updated to include additional information on matrix batch correction and normalization.
Raw data for 16 new projects now available
April 12, 2021
Raw sequencing data for 16 new projects are now available in the HCA Data Portal Data Browser. These projects include single-cell data derived from:
- Human and mouse
- 10x 3’, 10x 5’, Smart-seq2 technologies
- Small intestine, aorta, brain, skeletal muscle, blood, pancreas, tonsil, lung, skin, immune system, kidney, and eye
- Airway basal stem cells exposed to SARS-CoV-2
- Disease states, including Crohn's Disease, aneurysm, Multiple Sclerosis, HIV, Type 2 Diabetes, and glioblastoma
Processed data now available for 26 HCA 10x datasets
April 02, 2021
The HCA Data Portal 2.0 Preview now has standardized BAMs and count matrices ( Loom file format) available for 26 HCA projects, including 15 new projects. These projects contain both human and mouse single-cell and single-nucleus data generated with 3' 10x V2 and V3 sequencing technology. This data was processed using the latest version of the Optimus pipeline.
In addition to individual sample count matrices, each newly processed project also has standardized, project-level HCA Data Portal-generated matrices that are stratified by organ, species, and library construction method. These matrices are minimally filtered to include only cells with more than 100 UMIs. You can download the project-level HCA Data Portal generated Matrices from the Data Browser (see image below) or from the individual Project page (see the Exploring Projects guide)
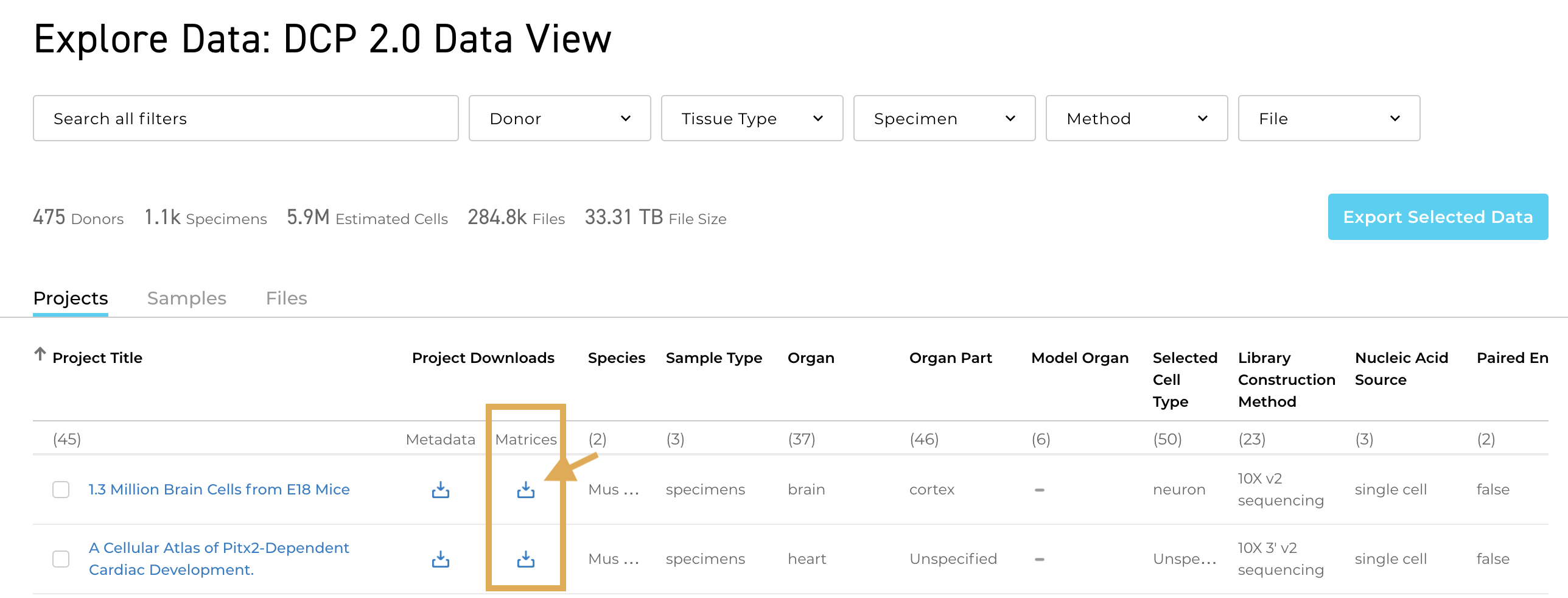
New raw data and Contributor-Generated Matrices
Raw sequence files and contributor-generated matrices (CGMs) for an additional 11 new projects are available for download from the HCA Data Portal 2.0 Preview. These projects include human adult, embryonic, and fetal data derived from 10 organs - brain, blood, bone marrow, colon, thymus, pancreas, placenta, mouth, liver, and spleen. Most projects employed 10x sequencing technology, but some additionally used cite-seq and drop-seq.
With the addition of these new data, the HCA Data Portal now has 55 total projects with over 7 million cells derived from 40 organs and 580 donors. We are grateful for the contributions and continued support of the 179 global labs who made this data available.
Download new data using updated guides
All files (raw data, CGMs, and HCA Data Portal-generated BAMs and matrices) can be downloaded following the instructions in the Accessing HCA Data and Metadata guide. Additionally, you can access matrix files programmatically using the new Programmatic Download guide.
HCA Data Portal 2.0 launches; new projects, contributor generated matrices and HCA Data Portal 2.0 infrastructure
December 11, 2020
In the spirit of bringing HCA data to the community as quickly as possible, we are releasing the new HCA Data Portal 2.0 data and infrastructure incrementally.
This initial launch -- the first of several planned to roll out new functionality, pipelines, and data -- includes:
- Raw data and standardized metadata for 5.8 million cells
- Contributor-generated matrices, embeddings, and annotations for existing and 16 new HCA Data Portal projects
- New HCA Data Portal 2.0 infrastructure (details below)
What's included in the new projects?
The new projects include a mix of human and mouse data from a variety of organs including adipose tissue, heart, hindlimb, spleen skin, yolk sac, diaphragm, tongue, trachea, and more. These data encompass:
- 143 donors
- 248 specimens
- estimated 1.3M additional cells
What's included in the HCA Data Portal 2.0 infrastructure updates?
Along with the new data view, we have updated HCA Data Portal infrastructure by retiring and replacing the following features:
- HCA Command Line Interface (CLI); data are now downloaded using curl commands (see the Accessing HCA Data and Metadata guide)
- HCA Matrix Service; HCA Data Portal 1.0 project matrices remain available on the individual Project Matrices page (see the Exploring Projects guide)
- HCA Data Storage Service (DSS) and API; data is now stored in the Terra Data Repo (TDR), an alternative storage and metadata management service that supports managed access
What's Coming Next?
Between now and the end of February look for:
- Standardized analysis results (BAM and index files) made by the latest HCA 10x and SmartSeq-2 pipelines for all projects
- HCA Data Portal-generated count matrices stratified by species, library construction method, and organ for each project
After these standardized analyses are complete, we will begin releasing new projects and analyses on a monthly basis. We are also working towards incorporating controlled-access projects.
Once processing is complete, we will retire HCA Data Portal 1.0 View.
Coming Soon - HCA Data Portal 2.0
October 9, 2020
We are excited to announce the launch of the HCA Data Portal 2.0 this fall. This has been a collaborative effort over the past 6 months to take on board the HCA community feedback and understand the unmet needs in order to align the goals of the HCA Data Portal with the wider HCA community goals.
We have made significant changes, including GA4GH-compliance, more data—50% more than previously—and, where available, contributor-generated count matrices, embeddings, and cell type annotations.
Changes Coming to Key Components
To accomplish this, we will be making the following changes and migrations:
- The current Data Storage Service (DSS) will be retired and we will migrate to the Terra Data Repo, which supports managed access, for storage and metadata management. Both Ingest and the Data Browser will access the Terra Data Repo via self-service APIs, making for minimal disruption to the HCA scientific community.
- The matrix service API will be retired. The per-project matrices that are most commonly used will be available in static form directly from the data browser.
- As a higher-level replacement for the DSS API, the Data Browser API is being prepared for use by the wider developer community and will be officially documented and supported by the HCA Data Portal team.
- The HCA-CLI will be retired and the Data Browser will provide a bulk download capability via curl (or similar) commands. Because the internal organization of the data store is subject to change with upcoming work on the metadata, direct calls against it are discouraged in favor of the new Data Browser API.
Transition Path to HCA Data Portal 2.0
More information about the transition to HCA Data Portal 2.0 will be announced in the coming weeks and we will endeavor to make this transition as seamless as possible to the HCA research and developer communities.
We will continue to integrate third-party portals and applications into the HCA ecosystem by linking from and integrating directly into the HCA Data Browser.
Regards,
The HCA Data Portal Team