The Human Cell Atlas: Overview of Metadata Structure
Table of Contents
Introduction
This document describes the structure of the HCA metadata standard as implemented and utilized by the Data Coordination Platform (DCP). More detailed specification of the format and syntax of the metadata schemas and their instantiation can be found in the metadata schema structure specification on Google Drive.
What is in this document?
- High-level overview of the schema structure
- Description of schema URL structure
Who should be reading this document?
- Data contributors
- Data consumers
- Members of external projects seeking alignment with HCA metadata standards
What isn't in this document?
- The set of principles specifically guiding the schema structure design
- How metadata standard is semantically versioned
Structure overview
Motivation
The primary motivations for having a structured HCA metadata standard are to:
- Enable data contributors to describe their project in a way that produces high-quality data that can be maximally exploited by consumers and developers
- Enable data consumers to interpret data efficiently and in a standardized way
- Support developers who rely on metadata to develop and implement analysis and visualization tools
- Adhere to FAIR data principles (paper)
The primary motivations for the entity model chosen to describe the HCA metadata standard are to:
- Handle transitions between biomaterial and file entities
- Enable independent versioning of schemas representing different entities
- Support modeling of future sample and experiment types without needing to drastically alter the current model
Metadata entity model
There are five major entities supported by the HCA metadata standard: Projects, Biomaterials (biological samples), Protocols, Processes, and Files.
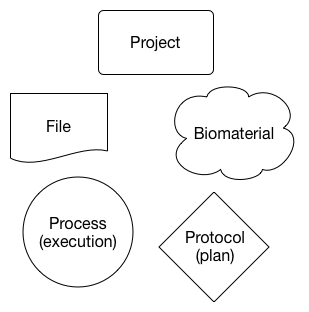
The entities are arranged in units that represent different parts of an experiment. For example, the diagram below is an abstract illustration of an input biomaterial (e.g., a tissue sample) undergoing a process (e.g., dissociation) to produce another biomaterial (e.g., a sample of dissociated cells). The process that was executed followed a specific protocol - or intended plan - to produce the output biomaterial.
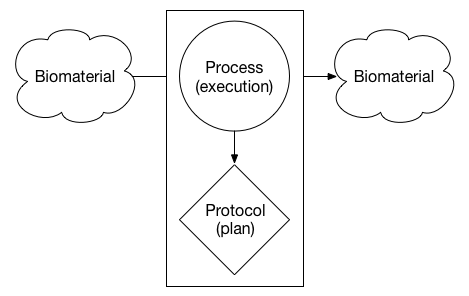
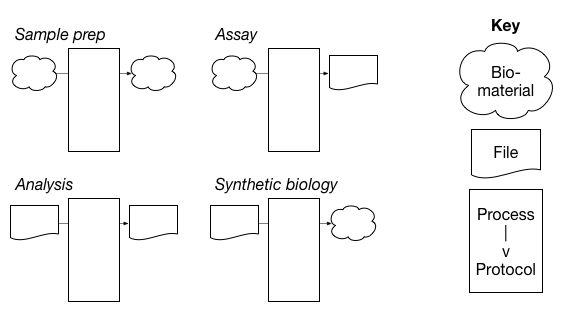
The metadata entity model supports units that can have one or more biomaterials or files as inputs or outputs. If both the input and output of a process are biomaterials, this unit represents some form of sample preparation. If the input is a biomaterial and the output is a file, the unit represents what many scientists refer to as an "assay". If both the input and output of a process are files, the unit represents an analysis. This flexible model allows for the possibility of modeling synthetic biology experiments - for example a file is used as an input to produce a custom biomaterial - in the future.
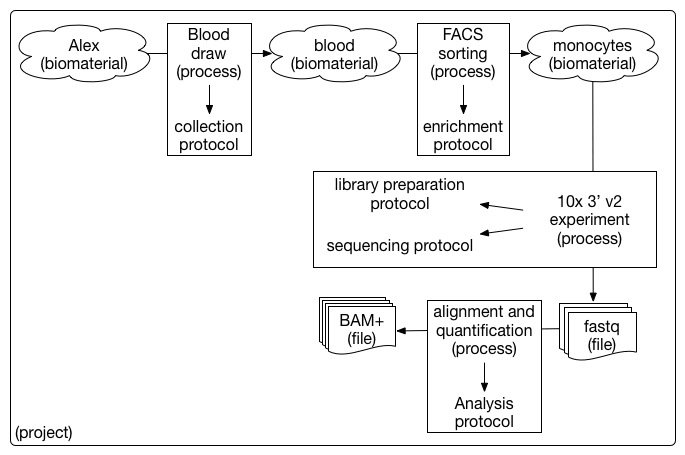
Below is an example single cell sequencing experiment modeled using the HCA metadata entity model.
Metadata field organization
Each instance of a project, biomaterial, protocol, process, and file entity is represented by a specific Type metadata schema. For example, in the HCA metadata standard each donor biomaterial is represented and described by the donor schema. Each Type schema includes a set of fields specific to describing that particular entity, and also inherits a set of core fields for that entity type - represented in a Core entity schema - and an optional set of thematically related fields - each set represented in a Module entity schema.
- Type entities = A specific instance of a project, biomaterial, protocol, process, or file entity. Type entities contain fields specific to that Type and inherit core fields from the corresponding Core entity.
- Core entities = Very stable, high-level entities that are referenced by Type entities. Core entities contain fields that apply to and are inherited by all corresponding Type entities.
- Module entities = Small, flexible entities that are extensions of an existing Type entity. Module entities contain extra fields related to a Type but that are thematically related.
User-supplied versus ingest-supplied metadata fields
Most metadata fields in Core, Type, and Module schemas are provided by data contributors during the submission process. A subset of fields in the metadata standard, however, are provided by the Ingestion Service component of the DCP. These ingest-supplied fields include the following:
provenancefield in all Type schemasdescribedBy,schema_version,schema_typefields in all schemas- all fields in
links.jsonandprovenance.jsonschemas
Ingest-supplied fields contain metadata about the submission (e.g. when metadata was uploaded to the DCP) and about the schema being used. Data contributors will never submit values to these fields directly.
In addition to ingest-supplied metadata fields, the ontology and ontology_label fields present in all ontology schemas will be populated by a look-up service based on the value supplied by data contributors in the corresponding text field in each ontology schema.
Recording the standard
The metadata standard is stored as a series of individual schemas which represent the entities and fields associated with them (e.g., project.json, biomaterial_core.json, sequencing_protocol.json). The schemas are stored in a single versioned control GitHub repository alongside documentation about the schema, the meaning of their content, and the update process. Anyone can propose changes to the schema through GitHub pull requests and issues. Only a specific list of committers will be allowed to approve pull requests and release new versions of the metadata schemas.
Specifying JSON schemas
Self-describing metadata schemas
Each JSON metadata schema is self-describing using the $id field with a URL to the location of that specific version of the schema.
In the donor_organism.json schema, the $id field looks like:
{
"$schema": "http://json-schema.org/draft-07/schema#",
"$id": "https://schema.humancellatlas.org/type/biomaterial/10.1.1/donor_organism",
...
}Self-describing metadata documents
Each metadata document for a Type entity requires a property - describedBy - that explicitly records the URI of the metadata schema which represents it.
In the donor_organism.json schema, the describedBy field is specified as:
"describedBy": {
"description": "The URL reference to the schema.",
"type": "string",
"pattern": "^(http|https)://schema.(.*?)humancellatlas.org/type/biomaterial/(([0-9]{1,}.[0-9]{1,}.[0-9]{1,})|([a-zA-Z]*?))/donor_organism"
}In a metadata document representing a Donor organism entity, the describedBy field is expressed as:
{
"describedBy": "http://schema.staging.data.humancellatlas.org/type/biomaterial/10.1.1/donor_organism",
...
}Schema URI structure
The structure of metadata schema URIs follows the convention:
http://schema.humancellatlas.org/{primary_directory}/{secondary/directory/structure}/{version}/{unqualified_schema_name}
where
{primary_directory}is one of [core,type,module]{secondary/directory/structure}describes the path to the schema, e.g.biomaterial,process/sequencing{version}is the version number of the schema, e.g.10.1.1{unqualified_schema_name}is the unqualified name of the schema, e.g.donor_organism
Some example URIs include:
http://schema.humancellatlas.org/core/biomaterial/5.0.1/biomaterial_core
http://schema.humancellatlas.org/type/biomaterial/5.0.0/cell_line
http://schema.humancellatlas.org/type/protocol/sequencing/5.0.0/library_preparation_protocol
http://schema.humancellatlas.org/module/ontology/5.0.0/cell_type_ontology
http://schema.humancellatlas.org/module/process/sequencing/5.2.0/barcode