Data Suitability
The first phase of the HCA will profile tens of millions of human cells, isolated and in their tissue context from major tissues of healthy research participants. This effort will combine many different types of cellular resolution data including single cell RNA-Seq, single nucleus profiling of frozen samples, and spatial analysis of cells in the context of tissues. Based on this, the HCA considers five criteria to assess the suitability of a dataset for building the Atlas now.
Currently, the HCA considers five criteria to assess the suitability of a dataset for building the Atlas.
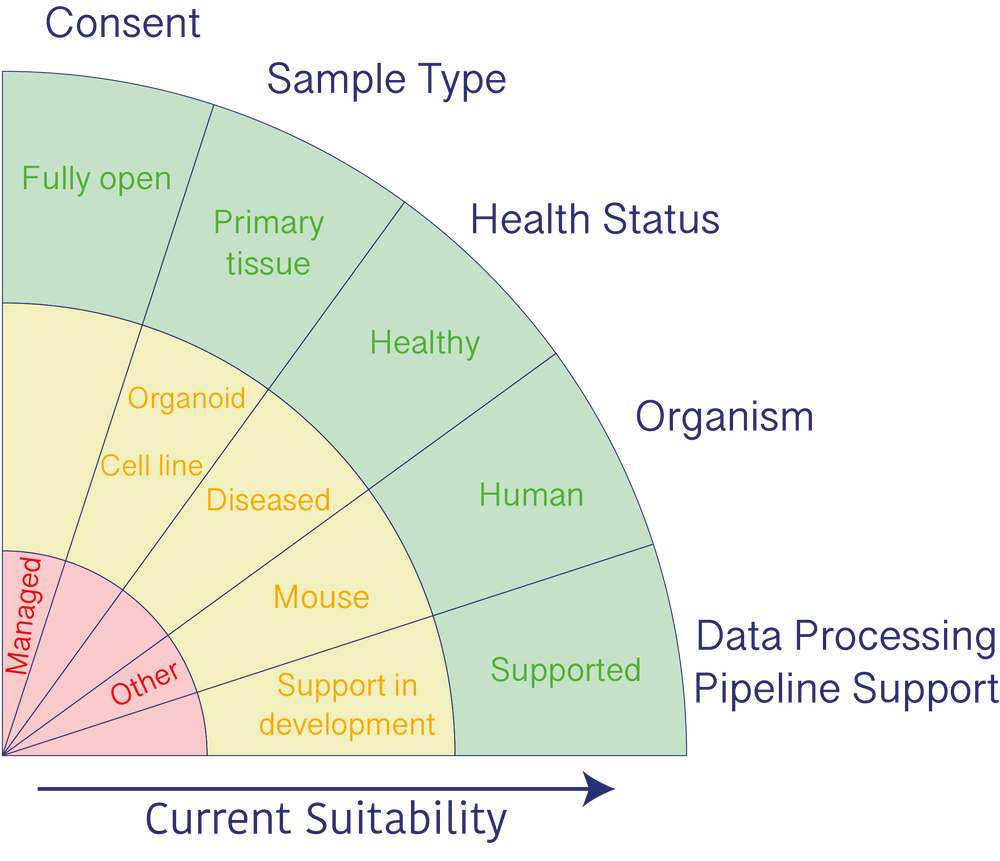
Each of the five criteria have the following statuses:
- Green: We are happy to receive your dataset.
- Amber: We are happy to discuss the value of your dataset to the HCA.
- Red: We are not collecting these data at the moment, but we would like to learn more about it.
Criteria
Consent
The current HCA data policy requires fully open access datasets. Any managed access data cannot be accepted at this time.
Sample Type
Current goals of the HCA focus on collecting data from primary tissues. For some organ systems or experimental approaches, primary tissue is not easily obtainable or openly consented. In these cases, we are happy to discuss accepting data from organoids, cell line models, or iPSC-derived cell cultures which provide representative biological systems.
Health Status
Currently, the HCA aims to define a baseline atlas of healthy cell types and states. The HCA recognizes that many groups work with samples from both healthy and diseased tissue. If consent allows, we are happy to accept a dataset that contains data from both healthy and diseased tissue, which may provide value to the Atlas.
Organism
The HCA intends to collect, process, and share data that comprehensively represents all human tissues. However, mouse models can provide access to some sample types and experimental strategies which are currently not possible in human. The current HCA data policy allows both human and mouse data to be submitted, but at this time, we are not accepting data from other species.
Data Processing Pipeline Support
The HCA currently supports processing of human 10x 3' v2 and paired-end Smart-seq2 scRNA-seq data. We are currently working to establish processing pipelines for droplet-based single nuclei RNA-seq, single end Smart-seq2 scRNA-seq, 10x 3' v3 scRNA-seq, support for mouse data for 10x 3' and Smart-seq2 scRNA-seq, and image-based transcriptomics data. We are happy to accept datasets and subsets of datasets that we cannot currently analyze to help us build and prioritize future pipelines.
Policy Changes
The HCA data suitability policy will evolve as required in response to new experimental and tissue preparation technologies, access to samples, and community requirements in order to build the HCA.
If you have data that meets any of the amber or red criteria and would like to discuss its suitability for the HCA, or if you have any further questions about this policy, please email wrangler-team@data.humancellatlas.org.
