Metadata
Overview
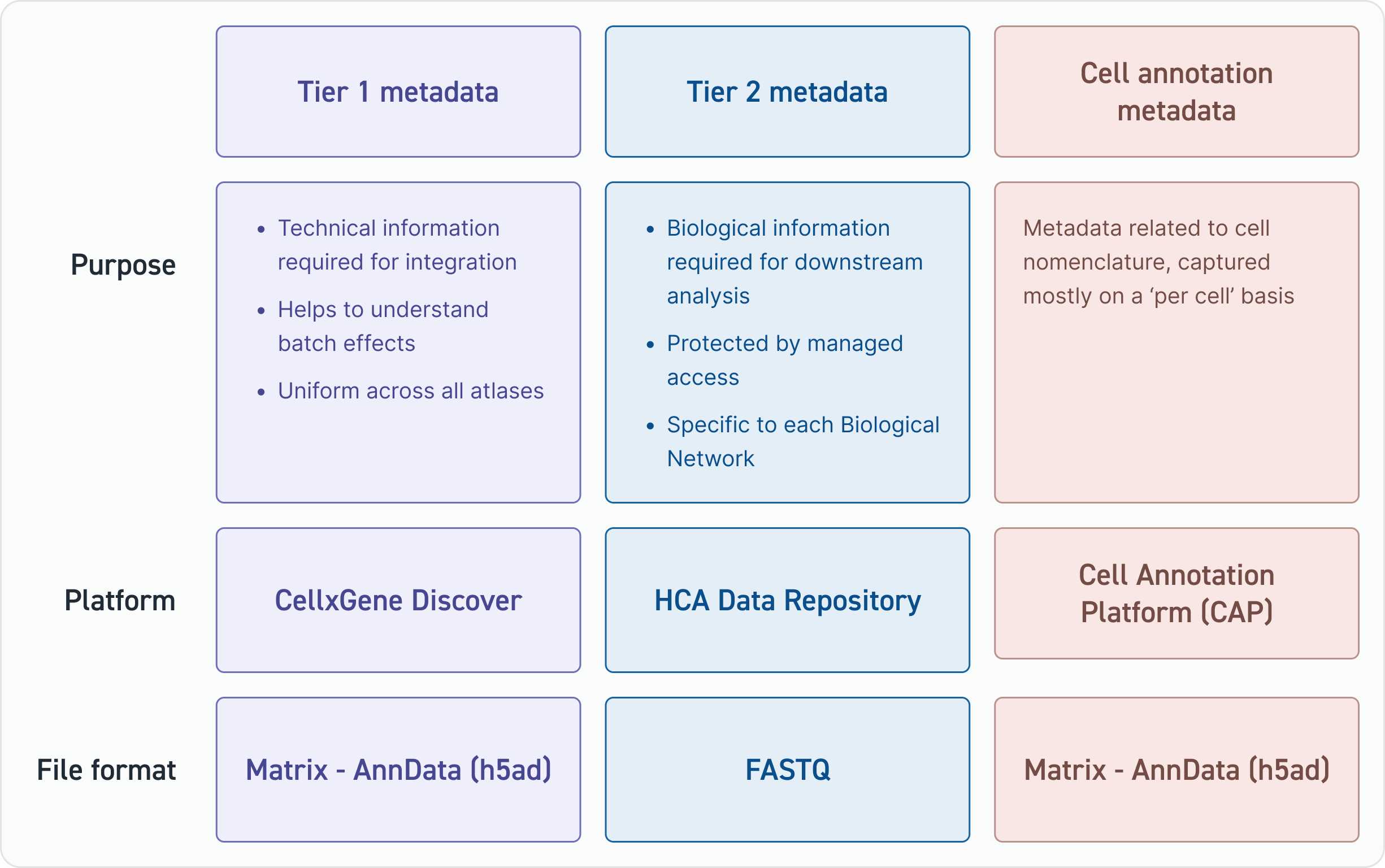
HCA has divided its metadata schema into three parts; each part serves a different purpose.
Tier 1 metadata fields are used to computationally integrate datasets, and hence, the fields are used to understand how successful the integration has been.
Tier 2 metadata files provide important information for scientists using HCA atlases in any sort of downstream analysis. These fields can contain identifiable information and, therefore, are stored under HCA’s managed access service. Finally, cell annotation metadata fields help to name cell types across all atlases.
Tier 1 metadata
Tier 1 metadata fields provide the foundational information used to build tissue and organ atlases. Tier 1 metadata fields are required to:
- Help identify samples or datasets included in the integration
- Help identify and explain technical batch effects
- Help qualify or disqualify datasets for inclusion in atlases
Understanding the factors that can cause batch effects is vital to ensure that when the datasets are combined into an atlas, they have not been over-corrected (i.e., masking true biological variation between cells) or under-corrected (e.g., resulting in the same cell types appearing as distinct from one another).
Submission of the Tier 1 fields is a prerequisite for atlas building and should be prioritized.
List of fields: here
Tier 2 metadata
There are two groups of fields that are captured under the umbrella of Tier 2 metadata:
- Fields that capture information that is not strictly required for the integration of datasets but is highly valuable for downstream biological analysis (these fields may differ per Biological Network)
- Fields requiring managed access protection (i.e., fields that contain potential identifiers)
These fields are separated from the Tier 1 fields for two reasons. First, these fields may take longer to wrangle, and we do not want to delay the teams building the integrated objects. Second, many of these fields require managed access protection for some data contributors and are therefore stored in a separate platform with this functionality.
The lists of fields are currently being defined by members of the Biological Networks. Completed fields lists will appear below once finalized.
- Adipose
- Breast
- Development
- Eye
- Genetic diversity
- Gut
- Heart
- Immune
- Kidney
- Liver
- Lung
- Musculoskeletal
- Nervous system
- Oral and craniofacial
- Organoid
- Pancreas
- Reproduction
- Skin
Cell annotation metadata
HCA’s cell annotation metadata fields relate to the naming of cells and will be stored on the Cell Annotation Platform (CAP).
CAP is a platform that allows individuals or groups to work on cell annotations in a private space before deciding to ‘publish’ the annotations for public viewing.
At the time of initial submission, a dataset does not need to have a finalized list of all cell annotations - that is one of the functions of CAP - it is a workspace to define cell annotations over time.
You will find that the cell annotation metadata requests detailed cell names (including ontology terms and synonyms), as well as the names of the overarching ‘parent’ populations (called ‘cell categories’). This helps to align cell names across datasets and create a hierarchy.
We highly recommend using the CAP user interface to input cell annotations, as it is easy to use and has helpful suggestions and prompts. Cell annotation fields are collected in order to:
- Find similar cell annotations between different cell annotation sets
- Define the similarities of the molecular signatures used to define individual “cell types”
- Create an “agreement/consensus” between cell annotations
List of fields: here
Amendments to the HCA schema
Over time, the HCA metadata schema has evolved to ensure metadata is insightful and inclusive of fields related to diversity. Datasets that have been integrated into atlases align to the newest version of the schema. Files submitted to the HCA Data Repository (previously known as the ‘Data Coordination Platform’ (DCP)) prior to mid-2024 will be aligned to an earlier version of the schema. The mapping between schema versions means that previously ingested data does not need to be amended.