Exploring Projects
Projects are a basic unit of data organization in the HCA Data Portal. Project contributors contribute raw sequencing and associated files along with rich metadata describing:
- the origin and type of the cells used in the project
- the processes and protocols used to collect and process the cells prior to sequencing
- the sequencing methods used
- details about the project contributors and their institutions
This Metadata is included in the project's Metadata Manifest (TSV file). When the HCA Data Portal processes the contributor's raw data with uniform pipelines, this processing information is also added to the Metadata Manifest.
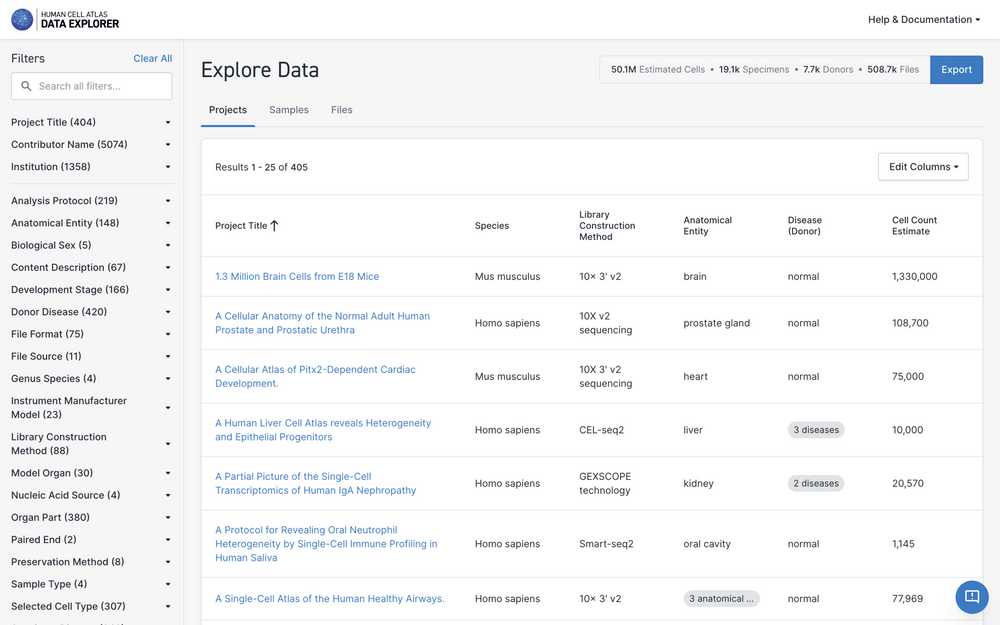
Finding a Project of Interest
The Data Portal Explore page lists all projects by title along with key project metadata. The project list is filterable by metadata values.
Viewing Project Details
Selecting a project title on the project list takes you to the project's information page.
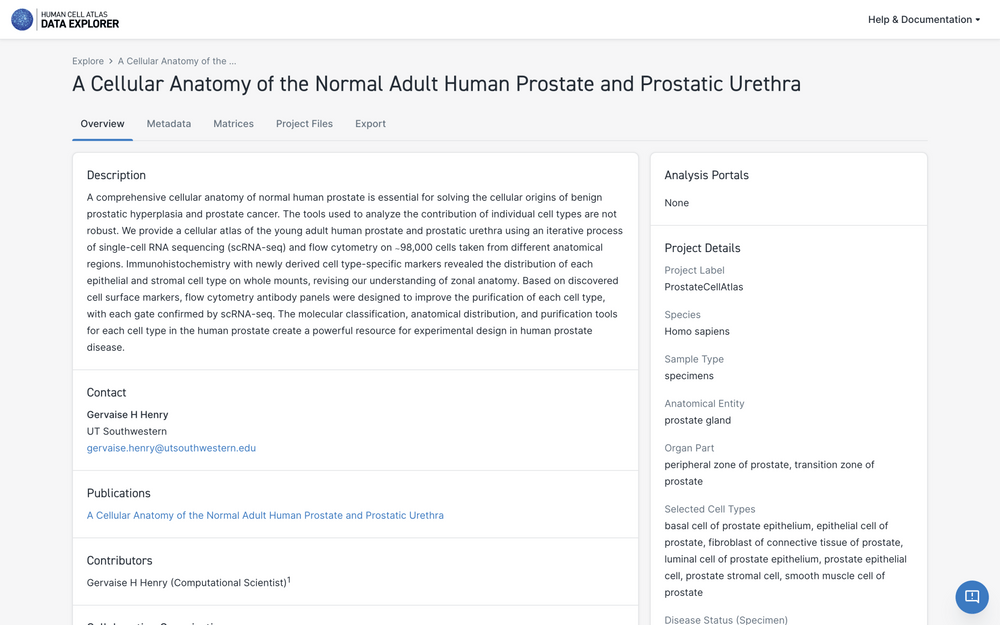
The project information page contains:
- the project title and description
- contributor information, collaborating organizations, and project contacts
- any publications or accessions associated with the project
- project details such as species, organ, and library construction method
- counts of input
- a link to the project metadata download
- a link to the project HCA Data Portal-generated count matrix download (if available)
- a link to the project contributor-generated matrix (if available)
Downloading Project Metadata
For each project, the HCA Data Portal maintains a project-specific TSV file containing the full project metadata. The TSV contains a row for each file in the project and columns for each metadata property. Meanings of the metadata properties are listed in the Metadata Dictionary.
The metadata TSV file gives a representation of the project's metadata graph that can be sorted and filtered using a standard spreadsheet or data manipulation tools.
The "Project Metadata" tab left of the Project Information page contains a link to download the project's metadata file.
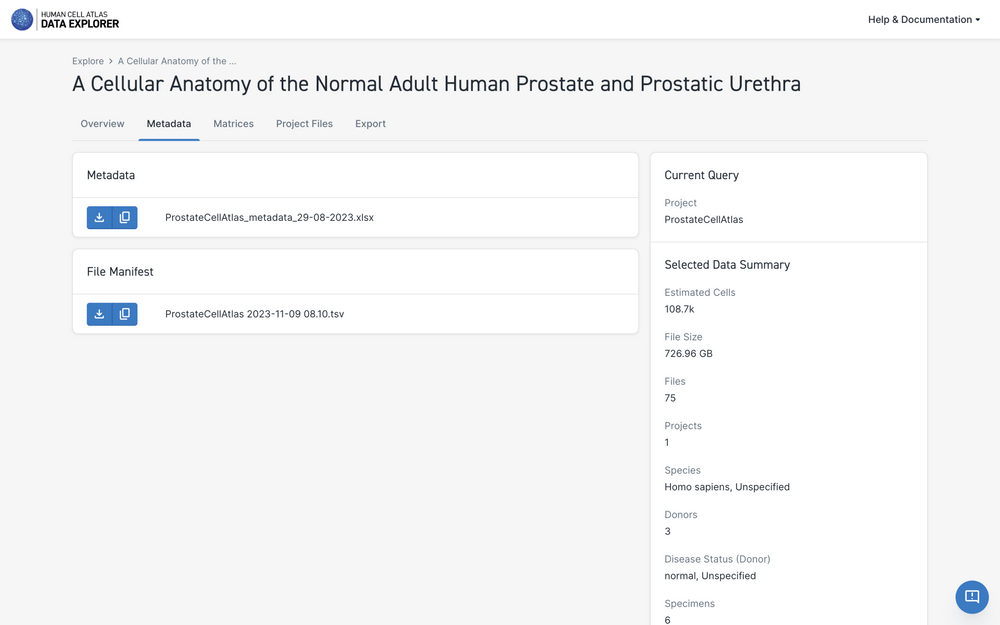
Metadata file sizes vary across projects but will generally be between 1 and 100 megabytes.
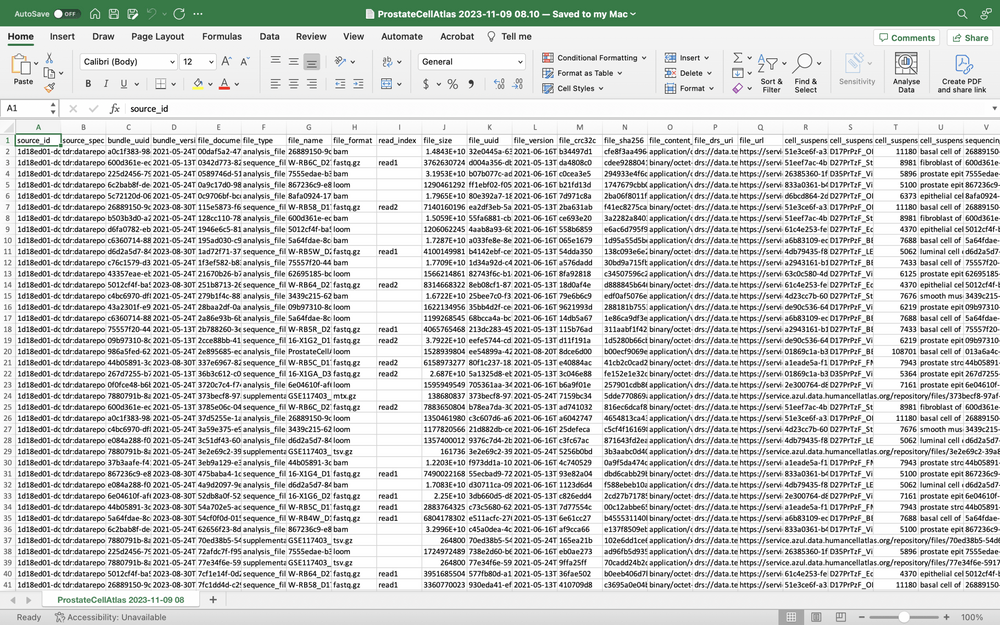
The TSV file is named after the project and includes the date and time the file was created. For example:
ProstateCellAtlas 2023-11-09 08.10.tsvA partial example of a TSV file is shown below:
Downloading Project HCA Data Portal-Generated Matrices
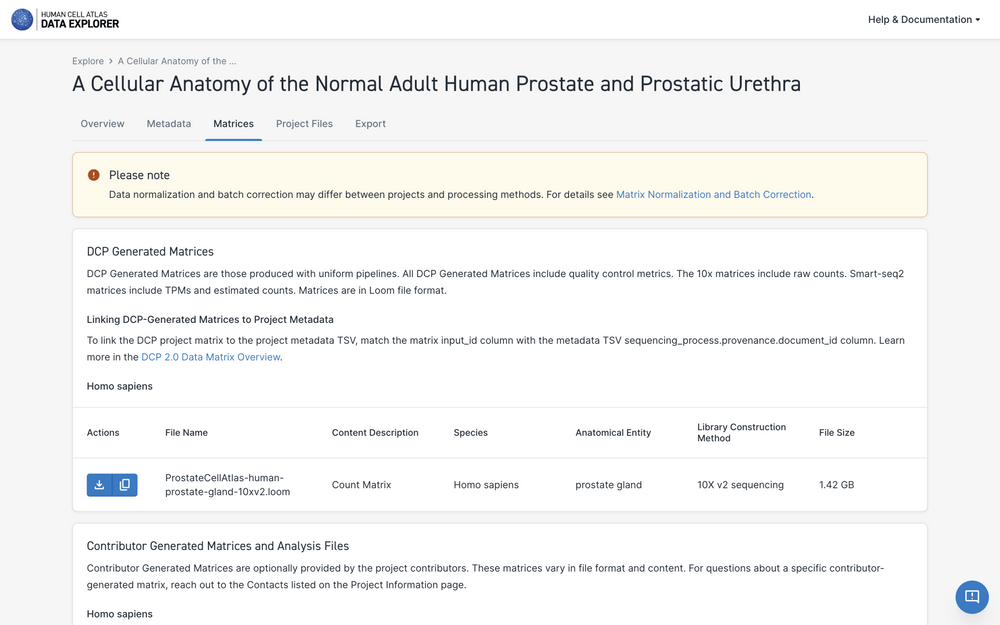
Each project processed with HCA Data Portal pipelines has HCA Data Portal-generated matrices. To download Project matrices, navigate to the Project Information page and select the "Project Matrices" tab to the left.
Scroll to identify the relevant matrix and then select the download icon.
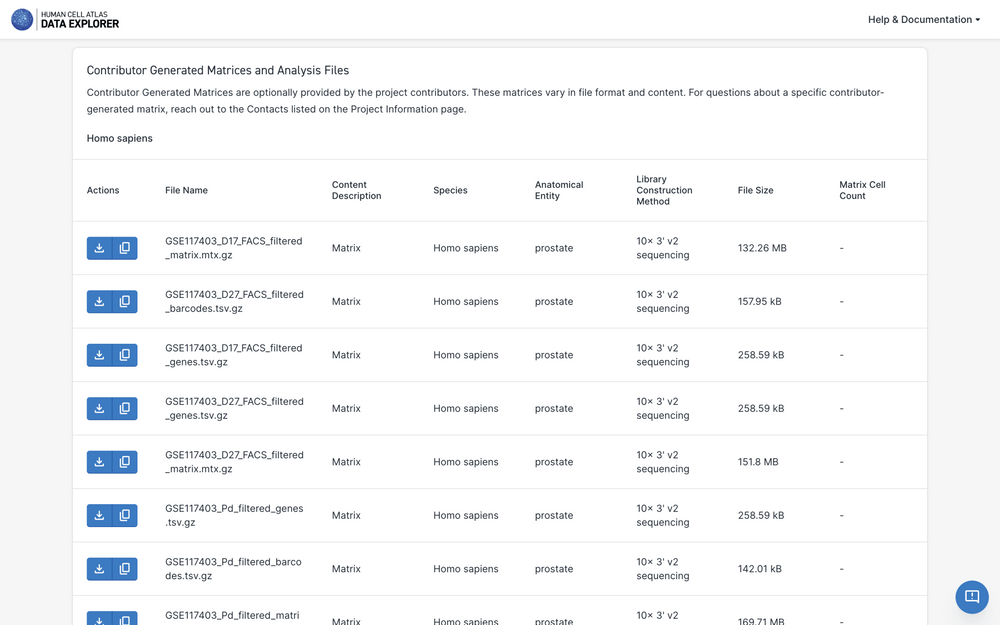
Downloading Project Contributor-Generated Matrices
Contributor-generated matrices are optionally provided by the project contributors. These matrices vary in file format and content. For questions about a specific contributor-generated matrix, reach out to the Project Contacts listed on the Project Information page.
To download the contributor-generated matrix, select the "Project Matrices" tab to the left of the Project page.
Scroll to the Contributor-Generated Matrices section and select the download icon.